Abstract
To assess the impact of the germinal center (GC) reaction on viral spread in Epstein–Barr virus (EBV) infection, we isolated EBV+ GC B cells from the tonsils of two infectious mononucleosis patients, sequenced their rearranged V genes, and determined expression of the EBV latency genes EBV nuclear antigen 2 and latent membrane protein 1. Most EBV+ GC B cells belonged to clones of cells harboring somatically mutated V gene rearrangements. Ongoing somatic hypermutation, the hallmark of the GC reaction, was seen only in uninfected GC B cell clones, not in EBV+ B cell clones. Thus, in infectious mononucleosis, GC and/or memory B cells are directly infected by EBV and expand without somatic hypermutation, whereas the GC passage of EBV-infected naive B cells does not contribute detectably to the generation of infected memory B cells, the main reservoir of EBV during persistence. Most, if not all, EBV-infected cells in GCs exhibited an unusual EBV gene expression pattern in that they were positive for EBV nuclear antigen 2 but negative for latent membrane protein 1. Although the three main types of EBV-associated B cell lymphomas (Burkitt's, Hodgkin's, and posttransplant lymphomas) presumably are derived from GC B cells, EBV+ GC B cells resembling these EBV+ GC B cell lymphomas in terms of EBV gene expression and somatic hypermutation pattern could not be identified.
Epstein–Barr virus (EBV) is a ubiquitous, tumorigenic herpes virus infecting human B cells (1). Whereas primary infection by EBV is usually asymptomatic during childhood, delayed primary infection manifests itself in about half of the cases in a disease called infectious mononucleosis (IM) (1). In both instances, a lifelong persistent infection is established in the B cell compartment in >95% of adults worldwide (1).
In latently infected B cells, distinct EBV gene expression patterns have been described. Besides expression of all latent EBV-encoded transcripts and proteins in latency III (also called growth program), more restricted viral latency gene expression patterns are observed under certain conditions (latencies I and II and latency program during persistence) (1). EBV-encoded genes differentially expressed in these latency programs were shown in vitro to mimic cellular proteins: EBV nuclear antigen (EBNA)-2 expressed in latency III is the main activator of viral and cellular genes and partially employs the Notch signaling pathways; latent membrane protein (LMP)-1 expressed in latencies II and III acts mainly like a constitutively active CD40 receptor; and LMP2A expressed in latencies II and III and perhaps in the latency program during persistence mimics and interferes with B cell antigen receptor signals (2–4).
Despite these insights, remarkably little is known about the effects of EBV infection on the cellular differentiation pathways of the infected cells in vivo. Likewise, the primary cellular target(s) of the virus in the B cell compartment is controversial. With respect to the latter, it has been suggested that EBV initially infects naive B cells, which subsequently undergo a germinal center (GC) reaction to gain access to the memory B cell pool, the main reservoir of EBV in healthy virus carriers (5). In the GC reaction, somatic mutations are introduced into V genes of antigen-activated B cells during proliferation. Subsequently, GC B cells are selected for high-affinity binding to the respective antigen and finally differentiate into memory B cells or plasma cells (6). Hence, this model proposed that EBV+ B cells behave in the GC like uninfected B cells.
However, our recent work on interfollicular EBV+ cells in IM tonsils suggested that the pool of infected memory B cells is generated by direct infection and subsequent expansion of these cells (7). Moreover, EBV-infected naive B cells may even be prevented from GC entry, as suggested by the blockade of GC formation in transgenic mice expressing LMP1 in B cells (8). These observations challenge the view that GC passage is a mandatory step in the general strategy of EBV to establish viral persistence.
To address these issues directly, we isolated EBV+ GC B cells from the tonsils of IM patients and analyzed their rearranged antibody V genes. This analysis allows identification of clonal expansions of B cells, because V gene rearrangements are unique for each newly generated B cell and are inherited by its descendants (6). Moreover, the origin of B cells and their participation in the GC reaction can be examined by the V gene sequence analysis, because somatic mutations are introduced into rearranged V genes in the course of the GC reaction. Thus, naive B cells carry unmutated V gene rearrangements and GC and memory B cells carry mutated ones, and B cell clones generated in the course of a GC reaction show intraclonal V gene diversity (9, 10). We also determined expression of EBV latent genes in such cells and, combining these results with those of the V gene analysis, attempted to identify putative nonmalignant progenitor cells for EBV-associated B cell malignancies, namely Burkitt's lymphoma (BL), Hodgkin's lymphoma, and posttransplant lymphomas, all of which are believed to be of GC B cell origin (1, 10).
Materials and Methods
Tissues.
In all seven cases of IM, tonsillectomy was performed because of complications that had developed during IM. Clinical data of six of these patients were published earlier (7). One additional patient, a 20-year-old man (patient 7), had been symptomatic for 2–3 weeks before tonsillectomy; recent EBV infection was confirmed by serology. Patients 1 and 2, studied in detail in the present analysis, were 16- and 17-year-old males who had been suffering for 4 days and 2–3 weeks from symptoms of IM, respectively, when tonsillectomy was performed.
Immunostaining, in Situ Hybridization, and Micromanipulation.
Immunostaining was performed on frozen sections as described (7) by using antibodies against EBNA2 (Biotest, Dreieich, Germany), CD3 (polyclonal rabbit anti-human CD3 antibody, DAKO), LMP1 (CS1–4, DAKO), CD20 (L26, DAKO), biotinylated secondary antibodies (DAKO and Santa Cruz Biotechnology), and diaminobenzidine as substrate for avidin-coupled horseradish peroxidase (Sigma-Aldrich, Deisenhofen, Germany, and DAKO, respectively). Fluorescent immunostaining was performed on frozen sections by using mouse antibodies against LMP1 and Bcl-6 (NeoMarkers, Lab Vision, Fremont, CA) and Alexa Fluor 594-conjugated goat anti-mouse IgG antibodies (Molecular Probes). Nuclear counterstaining was performed by using Hoechst 33258 (Sigma-Aldrich).
To screen IM tonsils for EBV-infected cells located in GCs, serial 7- to 10-μm-thick sections were cut, and EBV-encoded small RNA (EBER) in situ hybridization was performed on every 10th section (7).
Single EBER+ cells and groups of three to five EBER− cells were micromanipulated from frozen tissue sections of IM tonsils as described (7, 9). Aliquots of TBS buffer (143 mM NaCl/10 mM Tris, pH 7–7.5) covering the sections during micromanipulation served as negative controls. In addition, for case 1, single CD20+ B cells and CD3+ T cells isolated from adjacent sections were used as controls.
Single Cell PCR and Sequence Analysis.
The whole genomic DNA of isolated cells and controls was preamplified (11). VH, Vλ, and Vκ gene rearrangements were amplified by seminested PCR as described (7, 11, 12) by using V gene family-specific framework region I primers and J segment-specific primers. These Ig gene PCRs were performed for all samples and controls of cases 1 and 2. EBER+ cells of case 2 were analyzed for the presence of V gene rearrangements only if they were found to be positive in the EBV-specific PCR (see below). To exclude from the analysis potential errors of Taq DNA polymerase introduced during preamplification, individual sequence variations of clonally related cells were counted as mutations only if obtained repeatedly from independent aliquots of the preamplification reaction.
All samples were analyzed by PCR for the presence of fragments of the EBV genome. For samples of case 1, a fragment of the EBNA1 gene was amplified (7), and samples of case 2 were analyzed for the presence of a fragment of the LF2 gene. Thereto, two LF2-specific primers (EBV-F, 5′-GAGCTCTCCACAACAATGTTCCCTG-3′; EBV-R1, 5′-CGGGGAATCCACAGACATCCGCTA-3′) were added to the primer mix of the first round of PCR for rearranged VH genes. The second round for the LF2 fragment was carried out in a separate reaction by using 1× PCR buffer (Sigma-Aldrich), 2 mM MgCl2, 200 μM each dNTP, 125 nM each primer (EBV-F and EBV-R2, 5′-GCTCCCCTGTGATTATTCCTCCTAA-3′), and 1.5 units of Taq DNA polymerase. The annealing temperature during the 35 cycles of the second round was 58°C. The ability to detect single EBV genomes by these EBV-specific PCRs was confirmed by analyzing single sorted cells of cell line Namalwa, described as carrying one to two copies of EBV genome per cell (13).
Gel-purified PCR products were sequenced on an ABI 377 sequencer (Applied Biosystems). DNASIS software (Hitachi, Tokyo) and the V BASE database (www.dnaplot.de/) were used for sequence analysis. V gene sequences have been submitted to GenBank (accession nos. AJ506390–AJ506648).
Results
Rare Occurrence of EBV-Infected Cells in GCs.
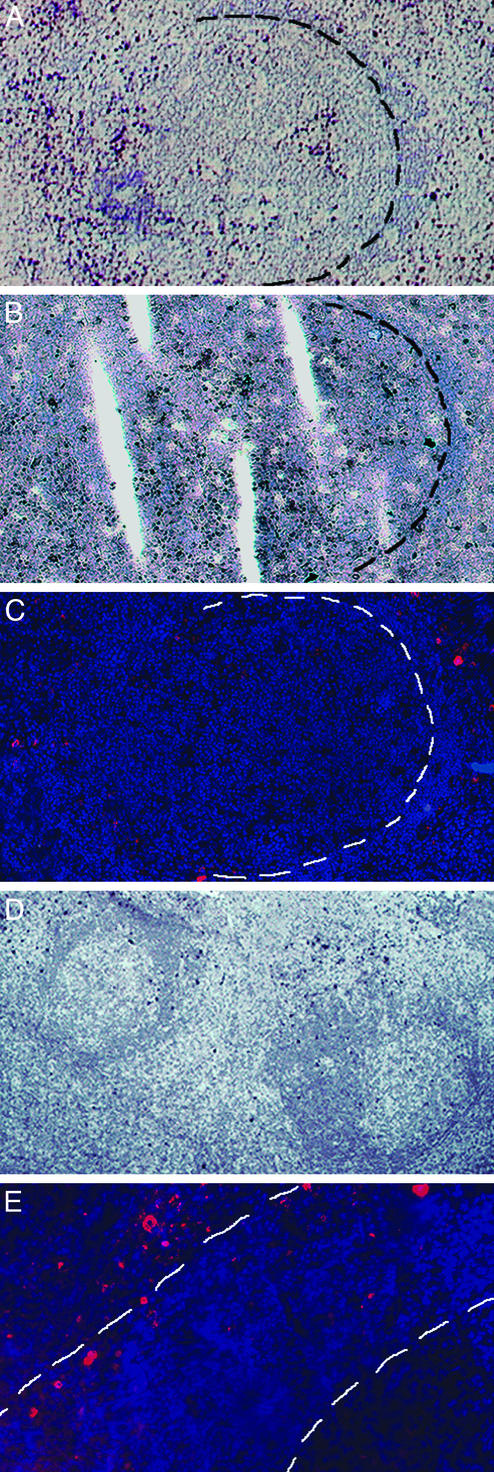
To screen for EBV-infected cells located in GCs during IM, sections of 20 tonsillar samples from seven patients suffering from IM were stained for EBER transcripts, which are expressed by all EBV-infected B cells. Most EBER+ cells were detected in the interfollicular region (IFR). Although a few GCs were found in sections of each case, substantial numbers of EBER+ cells were observed in GCs in only two tissue samples. In case 1, many EBER+ cells (>100 per section) were found in, or closely associated with, a single GC whose mantle zone seemed to be partly disrupted (GC1, Fig. 1A). In case 2, some EBER+ cells were located in two neighboring GCs (10–20 cells per GC per section) and in the IFR between them (GC2 and GC3, Fig. 1D).
Figure 1.
Phenotypic characterization of EBV-infected cells located in GCs and the IFR. Staining was performed on frozen tonsillar sections. Counterstaining was performed by using haemalaun (blue staining, A, B, and D) or Hoechst 33258 (blue fluorescent staining, C and E). (A–C) Sections of case 1 stained for EBER transcripts (dark blue-purple staining, A) and EBNA2 (dark brown staining, B) and LMP1 expression (red fluorescent staining, C). The border of GC1 to its mantle is indicated. (D) Section of case 2 stained for EBER transcripts (dark blue-purple staining). Approximately 10 EBER+ cells are located in GC2 (left) and GC3 (right). (E) Section of case 2 stained for LMP1 expression (red fluorescent staining) showing the interfollicular location of LMP1-expressing cells. Borders of GC3 and its mantle zone are indicated. A part of GC3 is visible in the lower right corner.
Expression Pattern of EBV-Encoded Genes.
In the two cases of IM in which EBV+ GC cells were observed, expression of the EBV-encoded proteins LMP1 and EBNA2 was determined for these cells and for EBV-infected cells located in the surrounding IFR. In both cases, cells strongly expressing LMP1 were detected in the IFR (Fig. 1 C and E). However, all three GCs (GC1–GC3) were essentially devoid of LMP1+ cells (Fig. 1 C and E). Only occasionally could a weak LMP1 signal be detected, in perhaps one or two cells per GC section. Compared with the number of EBER+ cells on adjacent sections, most, if not all, EBV+ cells express EBNA2 in GCs and the surrounding IFR (Fig. 1B). Hence, most EBV-harboring cells outside GCs of IM tonsils are positive for EBNA2 and/or LMP1, whereas most, if not all, EBV-infected cells in GCs express EBNA2 but lack expression of LMP1.
PCR Analysis of EBV-Infected Cells Located in GCs.
For the characterization of the population of EBV+ cells located in GCs, in terms of stage of differentiation and clonality and of their relationship to EBV+ cells outside GCs, single EBER+ cells from GCs and the IFR were isolated by micromanipulation and analyzed by PCR for Ig gene rearrangements (Table 1). To confirm that the structures analyzed represent functional GCs, groups of three to five EBER− GC cells for all three GCs were analyzed for rearranged V genes (Table 1). In addition, in case 2, groups of three to five EBER− mantle zone cells from GC2 and GC3 were analyzed. Performing EBV genome-specific PCRs able to detect single EBV genomes allowed confirmation of EBV infection of isolated EBER+ cells, and contamination of samples of EBER− cells by EBV-infected cells was excluded for the vast majority of samples (Table 1). Aliquots of buffer covering the sections during micromanipulation and single T cells isolated from adjacent sections of case 1 served as negative controls.
Table 1.
Sequence analysis of Ig gene rearrangements
| Case | Cell type | Location | No. of EBV PCR-positive samples/samples analyzed* | No. of V genes
|
% of cells (rearrangements†) with mutated V genes‡ | % of cells (rearrangements†) assigned to clones | Clones with mutated V genes/total no. of clones | Intraclonal V gene diversity | ||
|---|---|---|---|---|---|---|---|---|---|---|
| VH | Vκ | Vλ | ||||||||
| 1 | EBER+ | GC1 | 30 /47 | 16 | 1 | 100 | 94 | 1 /1 | ||
| IFR§ | 47 /68 | 18 | 100 | 100 | 1 /1 | |||||
| All | 77 /115 | 34 | 1 | 100 | 97 | 1 /1¶ | No | |||
| 2 | GC2 | 11 /32 | 4 | 4 | 4 | 84 | 67 | 3 /3 | ||
| GC3 | 17 /28 | 7 | 11 | 5 | 100 | 73 | 6 /6 | |||
| IFR§ | 114 /180 | 59 | 40 | 34 | 95 | 63 | 15 /15 | |||
| All | 142 /240 | 70 | 55 | 43 | 95 | 64∥ | 17 /17¶ | No | ||
| 1 | EBER−† | GC1 | 0 /45 | 13 | 16 | 11 | 66 | 20 | 3 /3 | Yes |
| 2 | GC2 | 0 /38 | 19 | 12 | 9 | 91 | 50∥ | 6 /6 | Yes | |
| GC3 | 3 /40 | 9 | 11 | 5 | 42 | 4 | 0 /1** | ? | ||
| Mantle of GC2 | 0 /22 | 14 | 12 | 12 | 10 | 0 | 0 /0 | |||
| Mantle of GC3 | 0 /16 | 8 | 10 | 7 | 0 | 8 | 0 /1 | No | ||
| Controls | ||||||||||
| 1 | B cells‡‡ | 11 /23 | 7 | 6 | 7 | 100 | 46 | 1 /1 | No | |
| T cells‡‡ | 4 /36 | 4†† | 1†† | 1†† | ||||||
| Buffer | 0 /41 | 0 | 0 | 0 | ||||||
| 2 | Buffer | 0 /59 | 0 | 0 | 0 | |||||
Samples of case 1 and case 2 were analyzed for the presence of a fragment of the EBNA1 and LF2 gene, respectively. For EBV+ cells the efficiencies of EBV-specific PCRs are expected to vary between 40% and 70% because of technical matters such as DNA instability and loss of cells during transfer into reaction tubes.
Because groups of three to five EBER− cells were isolated, numbers given for these samples refer to the number of rearrangements amplified.
Disregarding nonfunctional Vκ gene rearrangements, which are usually inactivated and hence exempted from somatic hypermutation.
Including some EBER+ cells located at the border of GCs. To avoid wrong assignment of these cells to the GCs, they were counted as cells from the IFR.
Members of individual clones were located inside, as well as outside, GCs.
From four members of two clones, unique V gene rearrangements were amplified that are incompatible with the clonal rearrangements obtained from other cells of these clones, indicating a low level of cellular contamination. These samples were not taken into further consideration.
For one clone consisting of a member isolated from GC2 and a member from GC3, only one member harbored somatic mutations in the clonal V gene rearrangement.
CD20+ B cells and CD3+ T cells were isolated from sections adjacent to sections from which EBER+ and EBER− cells were micromanipulated.
The four VH and the two Vλ gene rearrangements originate from four T cell samples. Although this indicates a low level of cellular contamination in the analysis, the reliability of the results is demonstrated by (i) the higher frequency of PCR products from EBER+ cells (30–43%) compared with the T cell samples (11%, data not shown), (ii) the lack of PCR products from 100 buffer controls, (iii) the very low frequency of EBV PCR products (2%) obtained from EBER− samples, and (iv) the fact that all V gene PCR-positive samples of EBER+ cells from case 1 were indeed positive in the EBV PCR.
EBV− B Cells Micromanipulated from GCs Participate in the GC Reaction.
From samples of EBV− GC cells isolated from GC1, GC2, and GC3, 40, 40, and 25 V gene rearrangements were amplified, respectively (Table 1). Disregarding nonfunctional Vκ gene rearrangements (14), 66%, 91%, and 42% of rearranged V genes were somatically mutated (Table 1) for GC1, GC2, and GC3, respectively. Because GCs are dynamic structures showing an increasing fraction of B cells with mutated V gene rearrangements during GC development (15), the high frequency of B cells with mutated V genes in GC1 and GC2 likely reflects an advanced stage of these GCs, whereas GC3 represents an earlier stage of development. In GC1, three clones with two members each were identified among rearranged V genes amplified from EBV− cells (Table 1). In case 2, 15 cells from GC2 can be assigned to six clones, and for GC3 one member of a clone was identified as related to a cell located in GC2 (see also Table 1 legend). Intraclonal V gene diversity, a hallmark of the GC reaction, was detected in eight of the nine clones identified among EBV−GC cells from GC1 and GC2 (Table 1). In contrast to GC B cells, the majority of EBER− mantle zone cells of GC2 and GC3 carried unmutated V gene rearrangements (90% and 100% of rearrangements, respectively; Table 1).
Taken together in GC1 and GC2, EBER− GC B cells are subject to ongoing somatic hypermutation, indicating that these cells undergo a classic GC reaction. In the case of GC3, histology, the expression of the GC-specific protein Bcl-6 (data not shown), and the presence of a mantle zone composed of B cells with unmutated V gene rearrangements also speak in favor of its GC identity. The low frequency of mutated and also clonally related cells suggests that this GC was at an early stage of development (9).
EBV-Infected B Cells Located in GCs Do Not Participate in the GC Reaction.
In case 1, rearranged VH genes were amplified from 16 and 18 EBER+ cells isolated from GC1 and the IFR, respectively. In addition, a Vκ gene rearrangement was obtained (Table 1) from one cell located in the GC. With the exception of one cell, all other 33 informative cells (97%) belonged to a single clone showing massive clonal expansion (Table 1). This clone is characterized by a mutated VH3 gene rearrangement (mutation frequency = 9.8%). None of the 33 members of this clone located in GC1 and the IFR showed intraclonal V gene sequence diversity (Table 1). Thus, in contrast to the neighboring EBV− cells, EBV-infected cells located in GC1 did not mutate their rearranged V genes during proliferation and hence did not participate in the normal GC reaction.
In case 2, 70 VH, 55 Vκ, and 43 Vλ gene rearrangements were obtained from 240 EBV-infected cells isolated from the two GCs and the IFR (Table 1). In GC2, GC3, and the IFR, 63–73% of informative EBV+ cells could be assigned to clones whose members were not restricted to one of the GCs or the IFR (Table 1). Altogether, members of 6 of 17 clones (with 2–16 members) were spread over the two GCs and the surrounding IFR. Whereas ≈5% of EBV-infected B cells carried unmutated V gene rearrangements (Table 1), all clonally expanded cells have mutated V gene rearrangements (mutation frequency for re-arranged VH genes = 7.8%). However, ongoing V gene hypermutation was not detected among EBV+ cells, despite ongoing somatic hypermutation in neighboring EBV− GC B cells (Table 1).
Thus, in both cases of IM analyzed, EBV+ cells located in GCs proliferate without ongoing somatic hypermutation, in striking contrast to the uninfected B cells in the same microenvironment. In addition, members of individual clones were spread over the GC(s) and the surrounding IFR.
Discussion
EBV-Infected B Cells Do Not Participate in the GC Reaction During IM.
In our previous analysis of EBV+ B cells in the IFR of IM tonsils, the detection of large clones of cells with somatically mutated V genes in the absence of ongoing somatic hypermutation suggested the establishment of the pool of EBV-infected memory B cells mainly by direct infection and subsequent expansion of such B cells (7). However, it has been proposed that the GC passage may be an important step in the virus's strategy to establish itself in the memory B cell compartment (5). Therefore, we wanted to clarify the impact of the GC passage of infected B cells for the spreading of EBV in the memory B cell compartment by direct molecular analysis of EBV+ GC B cells in IM tonsils.
In accordance with the literature and our previous study (7, 16–18), we show here that EBV-infected cells can be detected only occasionally in GCs of IM tonsils. The V gene analysis of the identified EBV+ B cells located in three GCs of IM tonsils demonstrates expansion of infected cells carrying somatically mutated V genes without ongoing somatic hypermutation. This argues against a recruitment of EBV+ naive B cells into the GC reaction and shows that EBV-infected GC or memory B cells do not participate in GC reactions even if they proliferate in the GC microenvironment. In addition, the present study shows spreading of members of EBV+ clones over the GCs and the IFR, indicating migration of EBV+ cells. Based on these results, two possibilities regarding the origin and differentiation of EBV+ B cells located in GCs of IM patients emerge. Thus, GC B cells may be infected by EBV and subsequently turn off somatic hypermutation while continuing to proliferate. Some members of the resulting clones of infected GC B cells may leave the GCs and migrate to the IFR or (as seen in case 2 of the present study) to other GCs. Alternatively, EBV may infect memory B cells that are driven to proliferate. Members of the resulting clones may enter GCs without taking part in the GC reaction. In the latter case, the observed proliferation may be driven by EBV infection rather than by antigens.
Taken together, these results support the concept that during IM a recruitment of EBV-infected naive cells into the GC reaction and subsequently into the memory compartment (5) does not play any significant role (7). Rather, the pool of EBV-infected memory B cells is established by direct infection of memory and/or GC B cells. The latter cells may differentiate into memory B cells without further participation in the GC reaction.
If this scenario also holds true for persistent infection, it would challenge the view that EBV transits from infected naive B cells into the pool of long-lived memory B cells via the GC reaction (5). However, it cannot be excluded on the basis of the present data that such a recruitment of EBV-infected naive cells contributes to virus persistence in healthy virus carriers, as proposed by Babcock et al. (5). In such individuals, this proposition would predict the rare occurrence of GCs in which a large fraction of B cells (descendants of one of the few GC founder cells, happening to be EBV-infected) is EBV-infected (and perhaps expresses LMP1) (5). Given that the frequency of EBV-infected B cells in healthy virus carriers is on the order of one in a million, this occurrence might have gone unnoticed in previous histological studies (17, 19).
EBV Gene Expression Patterns.
For the two IM tonsils analyzed at the molecular level, the expression pattern of EBV-encoded proteins LMP1 and EBNA2 was determined. EBV-infected cells in the IFR may express EBNA2 and/or LMP1 and thus may be composed of cells expressing distinct latency programs as suggested by previous studies (7, 16). However, in GCs, a more restricted pattern of EBV gene expression is observed: Most, if not all, EBV+ cells located in GCs express EBNA2 but lack expression of LMP1. This absence of LMP1+ GC B cells is in line with results from B cell-specific LMP1 transgenic mice (8), implicating incompatibility of LMP1 expression with entry of the infected cells into the GC microenvironment.
The finding that nearly all EBV+ cells located in the three GCs analyzed here express EBNA2 but lack expression of LMP1 is remarkable, because in classic EBV gene latency programs, EBNA2 expression always is associated with LMP1 expression. Only in rare cases of BL, AIDS-associated lymphomas, and posttransplant lymphomas do some tumor cells seem to express EBNA2 in the absence of LMP1 (20–22). Niedobitek et al. (16) also observed EBNA2+/LMP1− cells in tonsils of IM patients and suggested that this gene expression pattern is transitory and characteristic for recently infected cells. However, it is unlikely that the bulk of the EBNA2+ cells detected here in three GCs, as well as in 50–60% of EBV+ cells in the IFR of five IM cases in a previous study (7), are recently infected cells, because a large fraction of these cells belongs to large expanded clones of EBV-infected B cells. Because EBNA2+/LMP1− cells represent a major fraction of EBV-infected cells in each of the five cases of IM analyzed here and in the previous study, it also seems unlikely that these cells are shifting from latency III to latency I and acquire their immunophenotype because of the higher stability of EBNA2, compared with LMP1, protein. Rather, these cells may exhibit a distinct EBV gene expression program that needs to be characterized in more detail in future work.
Interference of EBV with Participation of Infected Cells in the GC Reaction.
The low frequency of EBV+ GC B cells in IM patients may have several causes. Thus, expression of LMP1 may interfere with the expansion of EBV+ B cells in GCs, because LMP1 mimics an activated CD40 receptor (2), and triggering of CD40 induces the differentiation of GC B cells to memory B cells that leave the GC (23). In addition, LMP1+ B cells residing in the IFR might be prevented from entering GCs, as inferred from the observation that LMP1 expression in murine B cells blocks GC formation (8).
Besides LMP1 expression, EBNA2 expression also may influence the behavior of EBV+ B cells in GCs. EBNA2 has been demonstrated to down-regulate Ig transcription (24), a prerequisite of somatic hypermutation. Therefore, EBNA2 expression may be responsible for the lack of hypermutation activity in proliferating EBV+ GC B cells. This idea is in line with several other observations: Whenever EBV-infected B cells were found to hypermutate in vivo (namely BL, B cell clones of angioimmunoblastic lymphadenopathy, and two recently investigated posttransplant lymphomas), the mutating EBV+ B cells did not express EBNA2 (refs. 25 and 26; A. Bräuninger, T. Spieker, A. Mottok, A. S. Baur, R.K., and M.-L.H., unpublished data). In contrast, ongoing somatic hypermutation was not observed in cases of posttransplant lymphoproliferative disease expressing EBNA2 in latency III (A. Bräuninger, T. Spieker, A. Mottok, A. S. Baur, R.K., and M.-L.H., unpublished data). The only instances reported so far in which EBNA2-expressing B cells undergo somatic hypermutation are two constitutively mutating BL cell lines (27). The reasons for this discrepant behavior remain to be explored.
While EBV-encoded genes thus may interfere with the GC reaction, they may well drive cellular expansion. Expression of EBNA2 and LMP1 or expression of EBNA2 together with CD40 triggering has been shown to cause proliferation of EBV-infected cells in vitro (28). Hence, in GCs, EBNA2 expression in conjunction with CD40 triggering may induce EBV+ cells to proliferate and to leave the GC microenvironment. Outside GCs, the expansion of the infected cells may be driven by the expression of EBNA2 in combination with LMP1.
EBV-Infected GC B Cells and Lymphomagenesis.
Given that the three main types of EBV-associated B cell lymphomas (BL, posttransplant lymphomas, and Hodgkin's lymphoma) presumably are derived from GC B cells (10, 29), we hoped that the phenotypic and molecular analysis of EBV+ GC B cells would shed some light on the relationship of the tumor cells to their nonmalignant putative counterparts. Therefore, we compared the features of these tumor cells with those of EBV+ GC B cells. However, neither ongoing somatic hypermutation, as often observed in BL and in some cases of posttransplant lymphomas (see above), nor crippling mutations (i.e., mutations rendering originally productive V genes nonfunctional) as described for tumor cells of Hodgkin's lymphoma (29) were detected in these cells. In addition, most, if not all, EBV+ cells located in GCs of IM tonsils exhibit a pattern of EBV gene expression (EBNA2+/LMP1−) differing from that seen in BL (EBNA2−/LMP1−), posttransplant lymphomas (EBNA2−/LMP1+ or EBNA2+/LMP1+) and Hodgkin's lymphoma (EBNA2−/LMP1+) (1). Hence, the EBV+ GC B cells present in IM patients do not resemble the tumor cells of the EBV-associated B cell lymphomas and their nonmalignant putative counterparts. The latter cells therefore either are rare among EBV+ GC B cells or develop in situations other than IM. In transplantation patients, for example, an impairment of T cell immunity may allow expression of immunogenic EBV-encoded proteins in a latency III program, resembling the EBV gene expression pattern seen in EBV+ posttransplant lymphomas and perhaps contributing to their transformation.
Acknowledgments
We thank Jochen Wustrow and John G. Strickler for providing material; Michaela Fahrig for technical assistance; Julia Jesdinsky and Mirela Stecki for sequencing work; Tilmann Spieker, Ines Schwering, Berit Jungnickel, and Ursula Zimber-Strobl for helpful discussion; Volker Diehl for support; and Martin Güldenagel and Manuel Montesinos-Rongen for help with graphic work. This work was supported by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 502 and a Heisenberg stipend (to R.K.).
Abbreviations
- EBV
Epstein–Barr virus
- IM
infectious mononucleosis
- EBNA
EBV nuclear antigen
- LMP
latent membrane protein
- GC
germinal center
- BL
Burkitt's lymphoma
- EBER
EBV-encoded small RNAs
- IFR
interfollicular region
Footnotes
References
- 1.Rickinson A B, Kieff E. In: Fields Virology. Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 2001. pp. 2575–2627. [Google Scholar]
- 2.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 4.Zimber-Strobl U, Strobl L J. Semin Cancer Biol. 2001;11:423–434. doi: 10.1006/scbi.2001.0409. [DOI] [PubMed] [Google Scholar]
- 5.Babcock G J, Hochberg D, Thorley-Lawson A D. Immunity. 2000;13:497–506. doi: 10.1016/s1074-7613(00)00049-2. [DOI] [PubMed] [Google Scholar]
- 6.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 7.Kurth J, Spieker T, Wustrow J, Strickler G J, Hansmann L M, Rajewsky K, Küppers R. Immunity. 2000;13:485–495. doi: 10.1016/s1074-7613(00)00048-0. [DOI] [PubMed] [Google Scholar]
- 8.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. Science. 1999;286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 9.Küppers R, Zhao M, Hansmann M L, Rajewsky K. EMBO J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küppers R, Klein U, Hansmann M L, Rajewsky K. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 11.Kanzler H, Küppers R, Helmes S, Wacker H-H, Chott A, Hansmann M-L, Rajewsky K. Blood. 2000;95:1023–1031. [PubMed] [Google Scholar]
- 12.Küppers R, Sonoki T, Satterwhite E, Gesk S, Harder L, Oscier D G, Tucker P W, Dyer M J, Siebert R. Leukemia. 2002;16:937–939. doi: 10.1038/sj.leu.2402480. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence J B, Villnave C A, Singer R H. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 14.Goossens T, Bräuninger A, Klein U, Küppers R, Rajewsky K. Eur J Immunol. 2001;31:3638–3648. doi: 10.1002/1521-4141(200112)31:12<3638::aid-immu3638>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 15.Weiss U, Zoebelein R, Rajewsky K. Eur J Immunol. 1992;22:511–517. doi: 10.1002/eji.1830220233. [DOI] [PubMed] [Google Scholar]
- 16.Niedobitek G, Agathanggelou A, Herbst H, Whitehead L, Wright D H, Young L S. J Pathol. 1997;182:151–159. doi: 10.1002/(SICI)1096-9896(199706)182:2<151::AID-PATH824>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Niedobitek G, Herbst H, Young L S, Brooks L, Masucci J, Crocker A B, Rickinson A B, Stein H. Blood. 1992;79:2520–2526. [PubMed] [Google Scholar]
- 18.Anagnostopoulos I, Hummel M, Kreschel C, Stein H. Blood. 1995;85:744–750. [PubMed] [Google Scholar]
- 19.Araujo I, Foss H-D, Hummel M, Anagnostopoulos I, Barbosa H S, Bittencourt A, Stein H. J Pathol. 1999;187:326–330. doi: 10.1002/(SICI)1096-9896(199902)187:3<326::AID-PATH242>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson C W, Niedobitek E, von Ostau C, Rooney N, Grasser F A, Young L S. Blood. 1997;90:1664–1672. [PubMed] [Google Scholar]
- 21.Hamilton-Dutoit S J, Rea D, Raphael M, Sandvej K, Delecluse H J, Gisselbrecht C, Marelle L, van Krieken H J, Pallesen G. Am J Pathol. 1993;143:1072–1085. [PMC free article] [PubMed] [Google Scholar]
- 22.Oudejans J J, Jiwa M, van den Brule A J, Grasser F A, Horstman A, Vos W, Kluin P M, van der Valk P, Walboomers J M, Meijer C J. Am J Pathol. 1995;147:923–933. [PMC free article] [PubMed] [Google Scholar]
- 23.Arpin C, Dechanet J, Van Kooten C, Merville P, Grouard G, Briere F, Banchereau J, Liu Y J. Science. 1995;268:720–722. doi: 10.1126/science.7537388. [DOI] [PubMed] [Google Scholar]
- 24.Jochner N, Eick D, Zimber-Strobl U, Pawlita M, Bornkamm G W, Kempkes B. EMBO J. 1996;15:375–382. [PMC free article] [PubMed] [Google Scholar]
- 25.Bräuninger A, Spieker T, Willenbrock K, Gaulard P, Wacker H H, Rajewsky K, Hansmann M L, Küppers R. J Exp Med. 2001;194:927–940. doi: 10.1084/jem.194.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman C J, Mockridge C I, Rowe M, Rickinson A B, Stevenson F K. Blood. 1995;85:2176–2181. [PubMed] [Google Scholar]
- 27.Harris R S, Croom-Carter D S, Rickinson A B, Neuberger M S. J Virol. 2001;75:10488–10492. doi: 10.1128/JVI.75.21.10488-10492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilger E, Kieser A, Baumann M, Hammerschmidt W. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanzler H, Küppers R, Hansmann M-L, Rajewsky K. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]