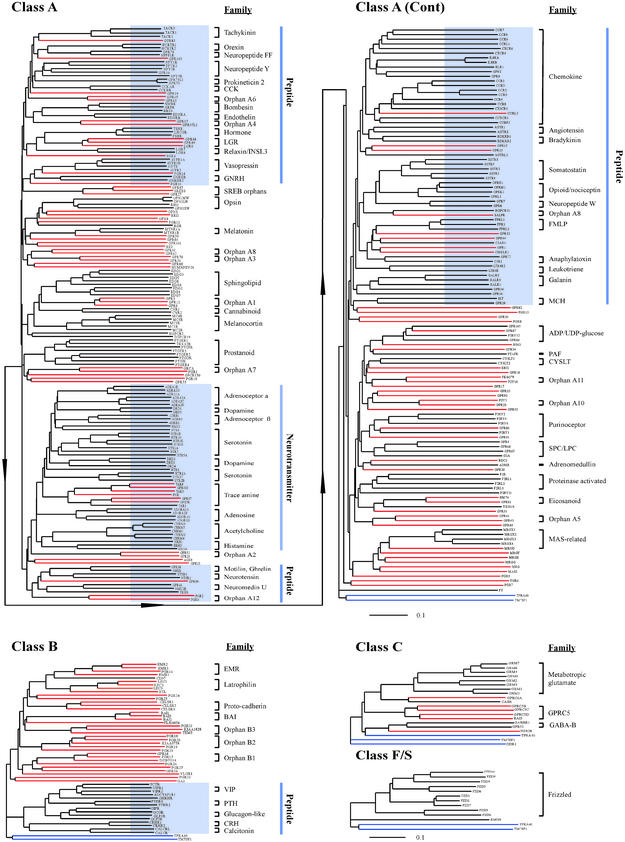
Figure 1.
Phylogenetic trees of human endoGPCRs. Lines corresponding to individual proteins are colored black for those with known ligands, red for orphan proteins, and blue for proteins with seven transmembrane domains but no homology to known endoGPCRs. The Class A tree was split in two parts due to size considerations (arrow line indicates the connection). Clusters of endoGPCRs with significant predictive value as to ligands are highlighted in blue on these bootstrap consensus trees (bootstrap values not shown). The ruler at the bottom of each tree indicates the horizontal distance equal to 10% sequence divergence.