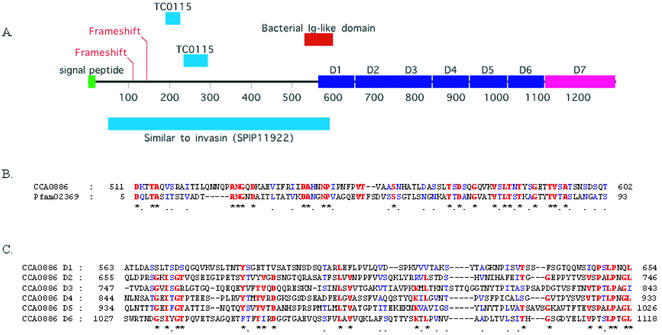
Figure 8.
Detail of the novel C.caviae invasin-like gene. (A) Schematic indicating relative location if the key domains within the invasin predicted protein sequence. The N-terminal half is most similar to Yersinia pseudotuberculosis invasin (SP:P11922) (72). A bacterial immunoglobulin-like domain also found in invasins and intimins is indicated. The C-terminal half shows no similarity to other invasins or intimins, lacking particularly several described sequences motifs and a disulfide loop required for binding to eukaryotic β1 chain integrins (73). Nonetheless, the presence of six repeats of approximately 90 amino acids (D1–D6) in this half suggests a similar structure and function in projecting a functional domain (D7, putative) away from the chlamydial outer membrane and into the milieu. The sequence encompassing the tandem repeats D2–D6 and some of the putative C-terminal functional domain D7 display sequence homology to the tandem repeat region of Bhp of Staphylococcus epidermidis, a paralog of Bap (74), involved in biofilm formation and virulence. The positions of the frameshifts in the nucleotide sequence and the location of the subsequences with similarity to C.muridarum TC0115 (see text) are also indicated. (B) Alignment of the bacterial immunoglobulin-like domain to the Pfam consensus sequence (75). Identical and conserved residues are indicated by red and blue, respectively. (C) Alignment of the intragenic repeats D1–D6 to each other. Identical and conserved residues are indicated by red and blue, respectively.