Figure 3.
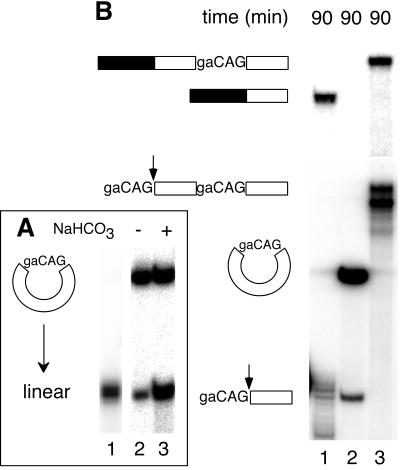
A circular 3′ substrate does not participate in bimolecular exon ligation. (A) Partial alkaline hydrolysis of the circular monomer. A single nick (lane 3) in the circular RNA (lane 2) first generates molecules of identical migration to linear RNA (lane 1); additional nicks produce a heterogeneous cleavage pattern beginning immediately below the full-length linear species. (B) Bimolecular exon ligation reactions (90 min) containing 175 nM linear monomer (lane 1), 175 nM circular monomer (lane 2), and 52 nM linear dimer (lane 3) 3′ substrates. Substrate and product structures are symbolized to the left. If the circular 3′ substrate had spliced, the ligated exon product would have been 5 nt longer than that from linear monomer. Note that the circular 3′ substrate was more stable in nuclear extract because it resists exonuclease digestion.