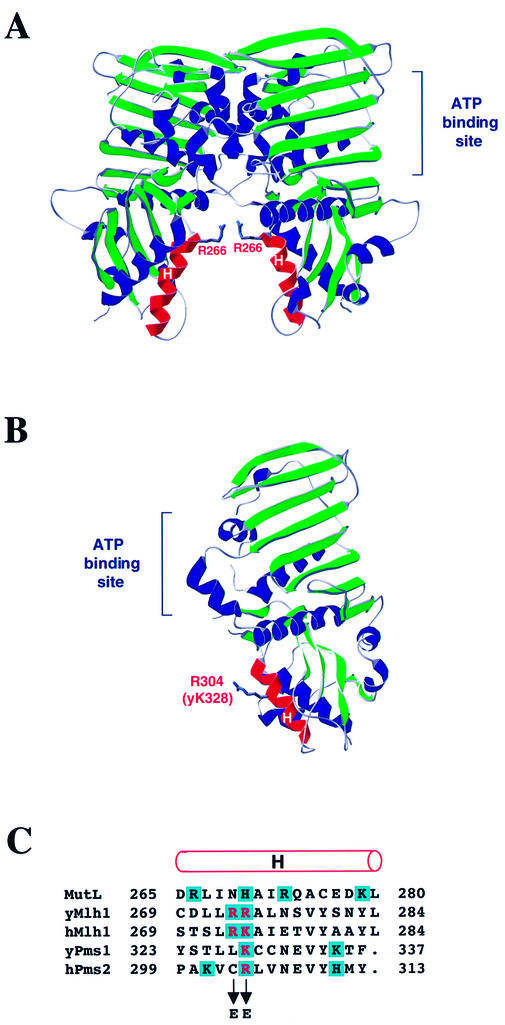
Figure 5.
Structure of MutL NTD and human PMS2 and alignment of MutL, MLH1 and PMS1. (A) Structure of the MutL NTD dimer (adapted from 19). The α-helices H in the proposed DNA binding groove between the two subunits are highlighted in red. The side chain of Arg266 is shown as sticks. (B) Structure of the human PMS2 NTD (adapted from 15). The α-helix H is highlighted in red. The side chain of the Arg304 residue homologous to Lys328 in yeast Pms1 and Arg274 in yeast Mlh1 is shown as sticks. (C) Alignment of amino acid sequences of E.coli MutL, S.cerevisiae and human MLH1, S.cerevisiae Pms1 and human PMS2 is according to Ban and Yang (14). The rod above the alignment designates α-helix H (19). Positively charged amino acid residues are shaded in blue. The residues that were changed to glutamic acid in this study are in red.