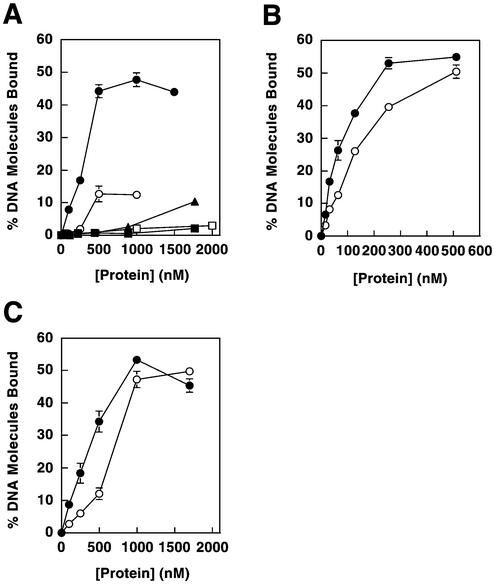
Figure 6.
Effects of Mlh1 mutations on DNA binding. (A) Amino acid replacements were made in Mlh1(343) and binding was determined for dsDNA and ssDNA substrates. Binding to pGBT9 is shown for Mlh1(343) wild-type protein (closed circles), Mlh1(343)-R274E (closed triangles) and Mlh1(343)-R273E/R274E (closed squares). Binding to poly(dT) is shown for Mlh1(343) wild-type (open circles) and Mlh1(343)-R273E/R274E (open squares). (B) Replacements in Mlh1 were incorporated into the Mlh1–Pms1 heterodimer. Binding to pGBT9 is shown for wild-type Mlh1–Pms1 (closed circles) and Mlh1-R273E/R274E–Pms1 (open circles). (C) Replacements were incorporated into full-length Mlh1. Binding to pGBT9 is shown for wild-type Mlh1 (closed circles) and Mlh1-R273E/R274E (open circles). All data represent the average of two to three independent experiments. Error bars show standard errors of the mean and are included for all data, but are not visible on some points because the error is smaller than the symbol itself.