FIG. 5.
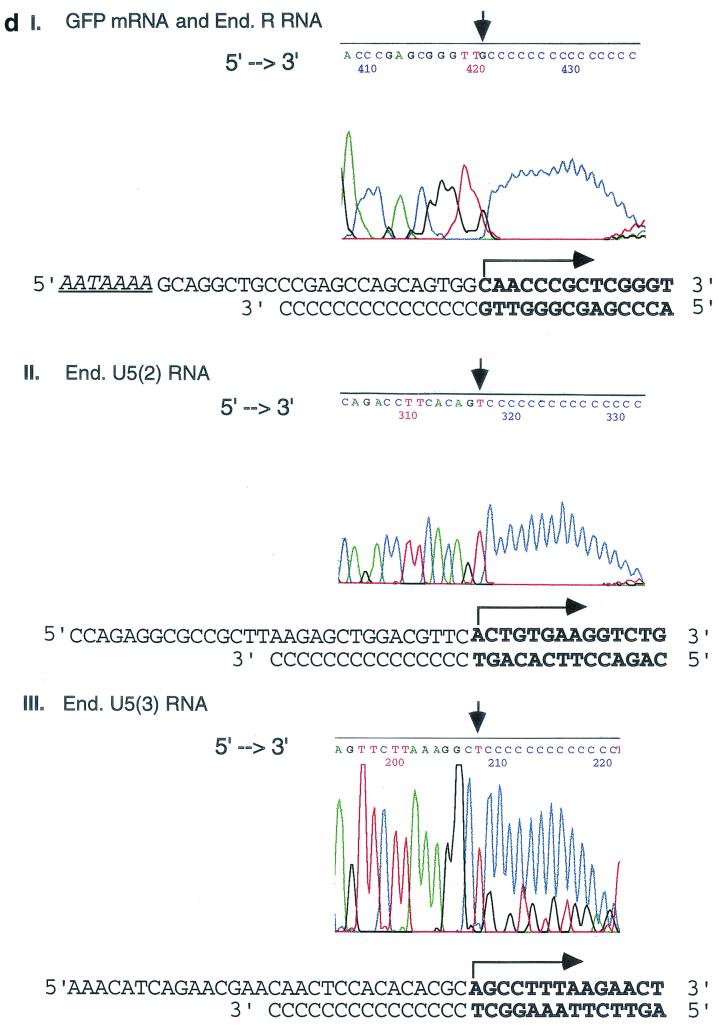
Transcriptional initiation sites of 5"HS5 LTR in transfected plasmids and in the endogenous human genomes mapped by 5"RACE. (a) Mapping the 5" ends of RNAs transcribed from 5"HS5 (E-P-R)-GFP plasmid integrated into K562 cells. Top, plasmid map. E, P, R, and GFP, same as in Fig. 1b. Angled arrow, transcriptional initiation site in the LTR marking the 5" border of the R region. Left-to-right arrows, GFP mRNA. Two right-to-left arrows, cDNA reverse transcribed from the GFP mRNA and DNA amplified from the cDNA template after 35 cycles of PCR, respectively. The PCR fragment depicted was the subsequently sequenced DNA strand; the arrows are aligned with the plasmid map on top. (C)n, poly(dC) tails added to the 3" ends of the cDNAs by the terminal deoxynucleotide transferase enzyme. Arrowheads at the 5" ends of the cDNA or the PCR fragment, reverse primers used for cDNA synthesis, PCR amplification, or DNA sequencing (seq.). Numbers, sizes in nucleotides of the cDNAs estimated from the locations of the initiation sites of the GFP mRNAs and of the PCR fragments determined from gel electrophoresis and DNA sequencing (see panels c and d). (b) Mapping of the 5" ends and initiation sites of the 5"HS5 LTR RNAs transcribed from the endogenous genome of K562 cells. Angled arrows, locations of the three transcriptional initiation sites mapped by 5"RACE; R with ∗, RNA initiation site found also in placental cells (see panel c, lane P); U5 with ∗∗, RNA initiation site found also in HeLa cells (see panel c, lane H). Left-to-right arrows, LTR R, U5(2), and U5(3) RNAs. The 5" ends of these three RNAs are located at the 5" borders of the R region and the second and third repeats of the U5 region, respectively. The 3" ends of the RNAs were drawn to the locations of the reverse primers used in the cDNA synthesis, although the actual 3" ends of the RNAs may be located further downstream. Right-to-left arrows and other designations, same as in panel a. (c) Gel electrophoresis of PCR fragments used for sequencing. The PCR fragments were generated from the following RNAs: Lane K1, GFP mRNA transcribed from the integrated 5"HS5 (E-P-r)-GFP plasmid; lanes K2, P, H, and K3, endogenous (End.) RNAs of nontransfected K562 cells, placental cells, and HeLa cells and a duplicate sample of the K562 RNAs, respectively. The band in lane K1 was generated by 35 cycles of PCR; the bands in lanes K2, P, H, and K3 were generated by 2 × 35 cycles of PCR with nested primers; and the 580-bp band in lane K3 was skewed upward due to a tear in the gel. Numbers on the right margins, sizes of the PCR DNAs in base pairs; lanes M, size markers; numbers on the left margins, sizes of the size marker bands in base pairs (the top three bands are 1,500, 1,250, and 1,000 bp, respectively; shorter bands are spaced 100 bp apart). (d) Electropherograms showing the locations of the 5" ends of GFP mRNA and the endogenous R RNA (panel I), the endogenous U5(2) RNA (panel II), and the U5(3) RNA (panel III). The electropherogram presented in panel I is generated by endogenous K562 5"HS5 LTR R RNA (the identical electropherograms of GFP mRNA and placental 5"HS5 LTR R RNA are not shown). 5"→3", the 5"→3" direction of the DNA sequences in the electropherograms. The vertical arrows mark the 3" ends of the cDNAs abutting the poly(dC) tails; the corresponding RNA initiation sites are marked with angled arrows in the DNA templates shown below the electropherograms. Boldface letters, transcribed bases in the sense DNA strand (top strands) and in the complementary cDNAs (bottom strands). For complete DNA sequences of the R and U5 regions, see reference 18.