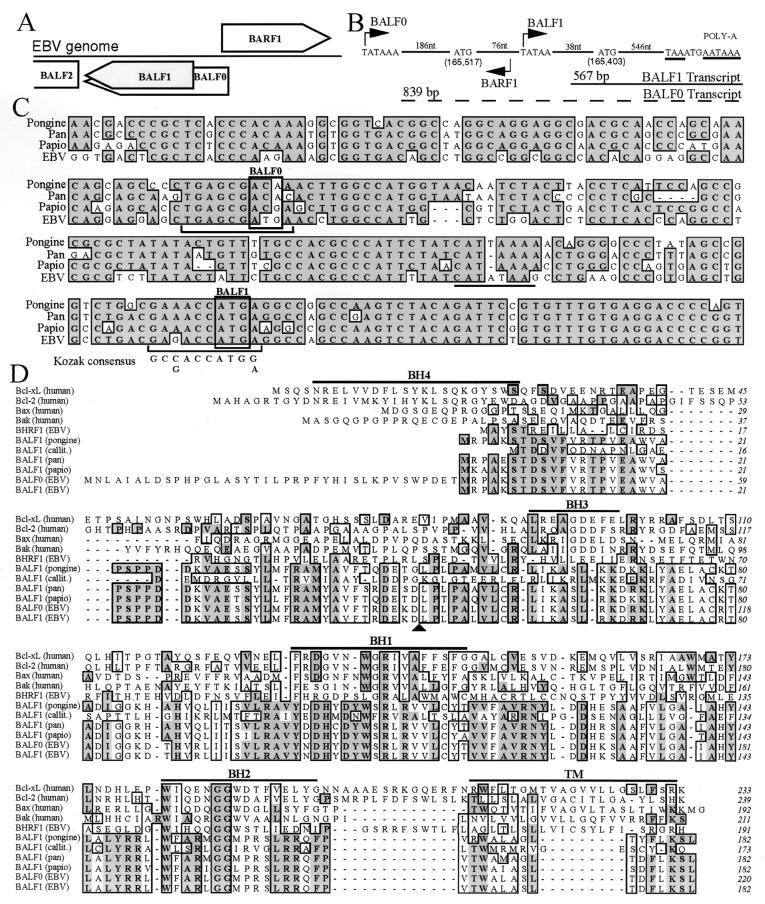
FIG. 1.
Human and primate herpesvirus BALF1. (A) Diagram of EBV genome region containing BALF1. Boxes that indicate the direction of transcription represent open reading frames. BALF1 (shaded) results from an internal initiation within the BALF0 reading frame. (B) Diagram of the EBV genome segment containing putative BALF1 transcripts (drawn in reverse of the usual orientation). Nucleotide sequences for putative TATA boxes (bent arrows), initiation codons, stop codon (underlined), and poly(A) signal (underlined) are shown. Numbers of intervening nucleotides (nt) are shown. Calculated transcript sizes [excluding poly(A)] are shown. (C) Alignment of the DNA sequences of BALF1 from EBV (V01555), pan herpesvirus (GenBank accession no. AF306944), herpesvirus papio (GenBank accession no. AF306943), and pongine herpesvirus 3 (GenBank accession no. AY034056). (D) ClustalW alignment of amino acid sequences of the indicated human and viral BCL-2 family members, including callitrichine herpesvirus 3 (GenBank accession no. AF319782). Identical (dark shading) and similar (light shading) amino acids occurring in 6 of 11 entries are marked. Homology domains BH1 to BH4 and the transmembrane domain (TM) of cellular BCL-xL are marked with horizontal lines. The BALF1 consensus caspase cleavage recognition site is marked by an arrowhead.