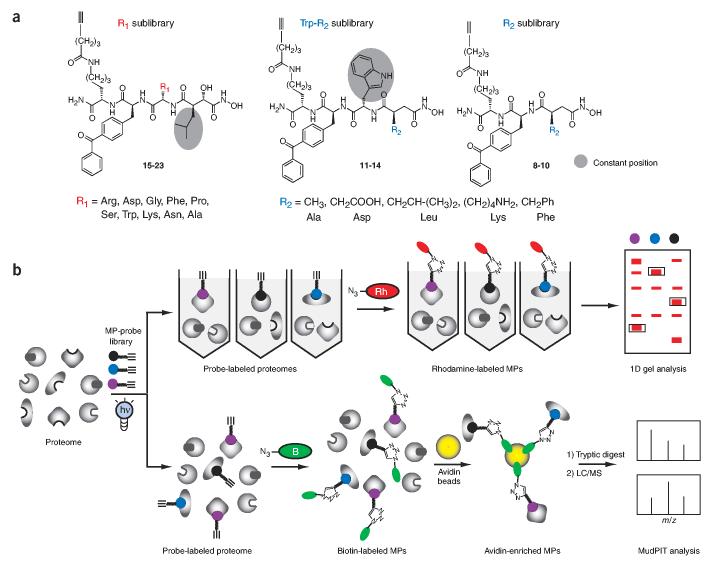
Figure 1.
Functional proteomic analysis of metalloprotease activities using active site-directed chemical probes. (a) General structure of the alkyne-tagged hydroxamate-benzophenone (HxBPyne) probe library for the proteome profiling of metalloprotease activities. The structures of the R1 and R2 substituents are shown and named based on the amino acid that they mimic (R1 sublibrary, 15-23; Trp-R2 sublibrary, 11-14; R2 sublibrary, 8-10). (b) Application of HxBPyne probes to proteomes and analysis of results by gel-based (upper) or MudPIT-based (lower) ABPP. In gel-based ABPP, individual HxBPyne-treated proteomes are reacted with an azide-rhodamine (Rh) reporter tag under CC conditions and separated by SDS-PAGE, and labeled metalloproteases are visualized by in-gel fluorescence scanning. In ABPP-MudPIT, proteomes are treated with a cocktail of HxBPyne probes and reacted with an azide-biotin (B) reporter tag under CC conditions. Probe-labeled metalloproteases are then captured on avidin beads, digested with trypsin and analyzed by multidimensional LC-MS.