Figure 1.
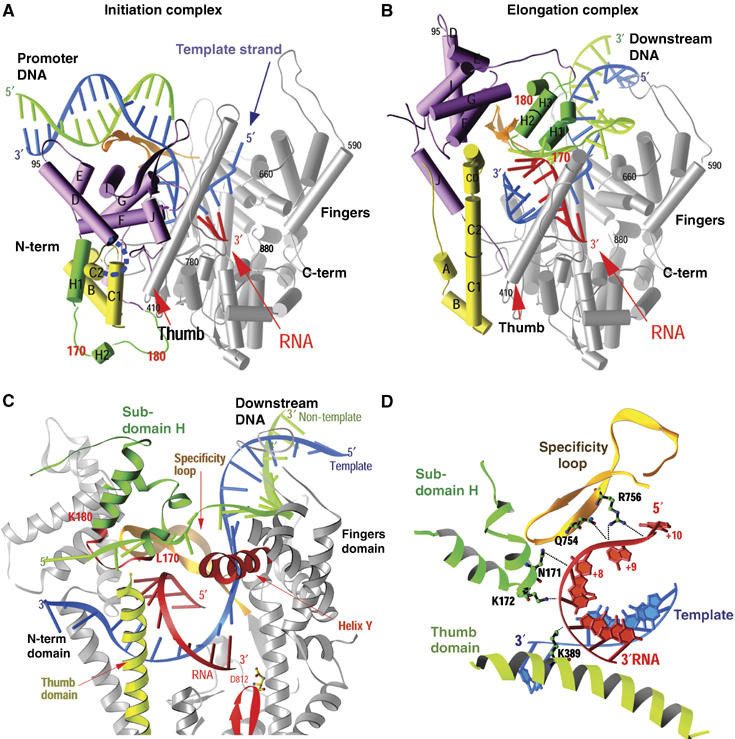
(A–D) Comparison of the structures of the T7 RNAP initiation and elongation complexes (A, B) and views of the transcription bubble (C, D) (Yin and Steitz, 2002). The initiation complex (A) and elongation complex (B) have been orientated equivalently by superimposing their palm domains. Helices are represented by cylinders and beta strands by arrows. The corresponding residues in the NH2-terminal domains of the two complexes that undergo major refolding are colored in yellow, green and purple, and the COOH-terminal domain (residues 300–883) is colored in gray. The template DNA (blue), non-template DNA (green) and RNA (red) are represented with ribbon backbones. The proteolysis-susceptible region (residues 170–180) is a part of subdomain H (green) in the elongation complex and has moved more than 70 Å from its location in the initiation complex. The specificity loop (brown) recognizes the promoter during initiation and contacts the 5′ end of RNA during elongation, whereas the intercalating hairpin (purple) opens the upstream end of the bubble in the initiation phase and is not involved in elongation. The large conformational change in the NH2-terminal region of T7 RNAP facilitates promoter clearance. This figure was made with the program Ribbons. (C) Interactions of the transcription bubble and heteroduplex in the elongation complex with domain H (green and red) and specificity loop (brown). Proteolytic cuts within the red loop in subdomain H reduce elongation synthesis (Ikeda and Richardson, 1987; Muller et al, 1988). Thumb sigma helix (yellow) and sigma helix Y (orange) are analogously involved in strand separation. (D) Side chains from subdomain H (green), the specificity loop (brown) and the thumb that interact with the single-stranded 5′ end of the RNA transcript and facilitate its separation from the template.
