Figure 6.
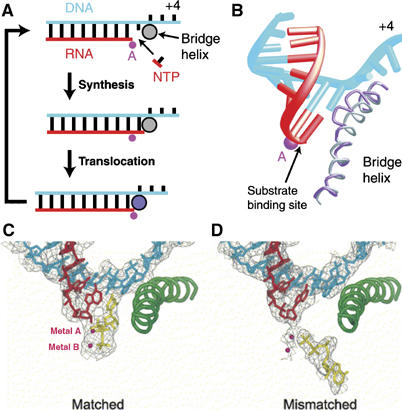
(A, B) Proposed transcription cycle and translocation mechanism (Gnatt et al, 2001). (A) Schematic representation of the nucleotide addition cycle. The NTP fills the open substrate site (top) and forms a phosphodiester bond at the active site (‘Synthesis'). This results in the state of the transcribing complex seen in the crystal structure (middle). Gnatt et al (2001) speculate that ‘Translocation' of the nucleic acids with respect to the active site (marked by a pink dot for metal A) involves a change of the bridge helix from a straight (silver circle) to a bent conformation (violet circle, bottom). Relaxation of the bridge helix back to a straight conformation without movement of the nucleic acids would result in an open substrate site 1 nucleotide downstream and would complete the cycle. (B) Different conformations of the bridge helix in Pol II and bacterial RNAP structures. The bacterial RNAP structure (Zhang et al, 1999) was superimposed on the Pol II transcribing complex by fitting residues around the active site. The resulting fit of the bridge helices of Pol II (silver) and the bacterial polymerase (violet) is shown. The bend in the bridge helix in the bacterial polymerase structure causes a clash of amino-acid side chains (extending from the backbone shown here) with the hybrid base pair at position +1. (C, D) Downstream end of the DNA–RNA hybrid in transcribing complex structures, showing occupancy of the A and E sites (Westover et al, 2004b). (C) Transcribing complex with matched NTP (UTP) in the A site. (D) Transcribing complex with mismatched NTP (ATP) in the E site. DNA is blue, RNA is red and NTPs are in yellow. Mg ions are shown as magenta spheres.
