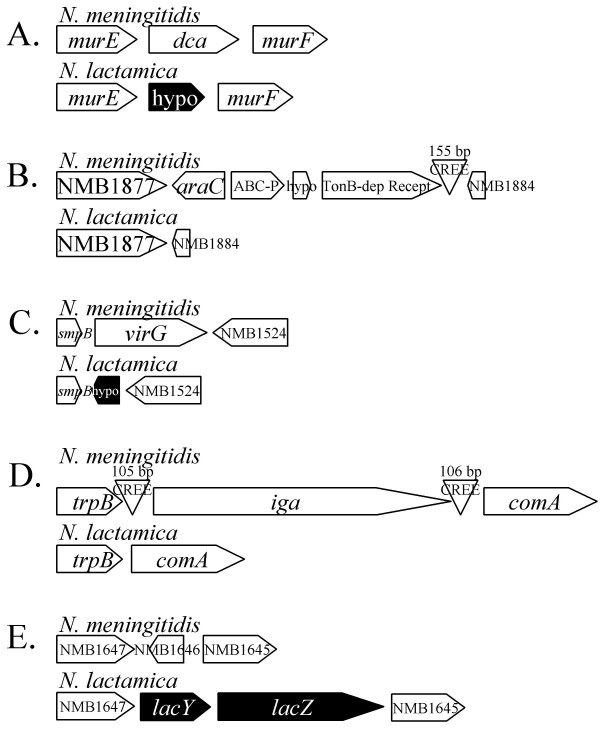
Figure 2.
The chromosomal locations of the 6 'virulence genes' present in all pathogenic Neisseria spp. genome sequences, but absent from the N. lactamica genome sequence and for which no N. lactamica strains hybridized to the pan-Neisseria microarray-v2. Panel A: the region between murE and murF contains dca in the pathogen genome sequences and a hypothetical gene in the N. lactamica genome sequence. Panel B: the region between NMB1877 and NMB1884 contains the 'virlence genes' NMB1880 (ABC-P, ABC transport periplasmic substrate-binding protein) and NMB1882 (TonB-dependent receptor) as well as a hypothetical gene (NMB1881) and an araC-like regulator in the pathogen genome sequences, which are all absent in this location in the N. lactamica genome sequence. The probe for NMB1878, the araC regulator hybridized to all N. lactamica strains assessed by CGH; this may be due to another araC-like regulator in another location or the presence of this regulator in this location in all the assessed strains. Panel C: the region between smpB and NMB1524 contains virG in the pathogen genome sequences and a hypothetical gene in the N. lactamica genome sequence. Panel D: the region between trpB and comA contains iga, flanked by Correia Repeat Enclosed Elements (CREE) commonly found in the neisserial genomes; iga and the CREEs are absent in the N. lactamica genome sequence. Panel E: the region between NMB1647 and NMB1645 contains a putative hemolysin encoded by NMB1646 in the pathogenic genome sequences, while in the N. lactamica genome sequence this is the location of lacY and lacZ, which confer the characteristic lactose fermentation phenotype of N. lactamica.