Abstract
A recognized hotspot for mutations conferring reduced echinocandin susceptibility (RES) is residue S645 of Candida albicans Gsc1(Fks1). We report that the mutation F641Y is associated with RES in a C. albicans isolate. The analogous Fks2 residue is mutated F to V in a Candida glabrata RES isolate; the introduction of this mutation into susceptible C. glabrata confirmed its role in RES. Y641-equivalent Fks residues were identified in intrinsically RES Fusarium species and Candida guilliermondii.
Echinocandins (ECs), such as caspofungin, are lipopeptide antibiotics that inhibit cell wall synthesis in susceptible fungi, which include yeasts and molds such as Candida albicans and Aspergillus fumigatus (1, 9). Their negligible toxicities, generally fungicidal activities, and lack of cross-resistance with azole antifungals have led to rapid clinical acceptance since their introduction in 2001. The EC target β-1,3-glucan synthase (GS) is membrane associated and hence difficult to study, but evidence points to the products of GSC1 in C. albicans or the related FKS1 in Saccharomyces cerevisiae as the major targets in these fungi. This evidence includes partial Gsc1 purification and correlation between gene dosage and GS activity (2). Moreover, multiple mutations that confer reduced echinocandin susceptibility (RES) have been mapped to GS genes. Specifically, three of four S. cerevisiae mutants selected on L-733560 (2), along with one of two mutants selected on Arborcandin C (13), involve an eight-amino-acid region of Fks1 (residues 639 to 646) or its homolog Fks2. Remarkably, C. albicans laboratory mutants selected on L-733560 (8, 14) and clinical isolates demonstrating reduced caspofungin (14) and micafungin (10) susceptibility have all been reported to have alterations in a single amino acid, S645, that falls within the equivalent eight-amino-acid region of Gsc1.
Candida glabrata has emerged as an important fungal pathogen in recent decades, most likely due to its intrinsic low-level and acquired high-level azole resistance (17, 20). Resistance following long-term treatment has threatened the use of azoles in C. albicans infections as well (22). ECs have equally high activities against azole-susceptible and -resistant strains of both of these species (the caspofungin MIC at which 90% of the tested isolates are inhibited was 0.06 μg/ml) (19) and hence provide a valuable alternative. ECs are, however, less active versus Candida guilliermondii and Candida parapsilosis (the caspofungin MIC at which 90% of the tested isolates are inhibited was 0.5 to 1 μg/ml) (16). Less susceptible still are Cryptococcus neoformans, Rhizopus oryzae, and Fusarium species (2). The basis for the intrinsic RES of these species is unknown. Not surprisingly, the expanding use of ECs has been accompanied by recent reports of RES in normally susceptible fungi (6, 7, 10, 12, 14, 15). Here we describe our characterization of two such clinical isolates, which identified novel mutations at equivalent positions within C. albicans and C. glabrata GS genes.
RES isolates and GS sequence analysis.
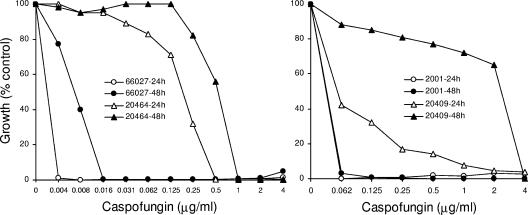
From a large collection of clinical isolates (16), single C. albicans and C. glabrata strains were selected that exhibited relatively high EC MICs when tested using the standard CLSI protocol with RPMI 1640 medium. Specifically, for C. albicans 20464, the caspofungin MICs were 1 and 2 μg/ml at 24 and 48 h, respectively, and for C. glabrata 20409, the MICs were 0.5 and 8 μg/ml at the same time points. To confirm and extend these data, these isolates, along with representative susceptible strains, were tested by broth microdilution assay in yeast extract-peptone-dextrose (YPD) medium, where the growth rate is higher and caspofungin MICs are typically lower than those obtained in RPMI 1640. C. albicans 20464 (Fig. 1) generated clearly defined MICs (≥80% inhibition) of 0.5 and 1 μg/ml after 24 and 48 h, respectively, of incubation while ATCC 66027 generated MICs ≥64-fold lower. Compared to C. glabrata strain ATCC 2001 (MIC, ≤0.06 μg/ml), C. glabrata 20409 appeared to be caspofungin susceptible at 24 h (Fig. 1); however, there was considerable trailing growth and, by 48 h, RES was pronounced with a caspofungin MIC of 4 μg/ml.
FIG. 1.
Broth microdilution assays examining caspofungin susceptibility of the indicated C. albicans or C. glabrata strains. Incubation was for 24 or 48 h, as indicated. Assays were performed in YPD medium at 35°C, as described previously (21).
For C. albicans 20464, a GSC1 region spanning the S645 codon mutational hotspot (14) was amplified and sequenced (see the Fig. 2 legend). A comparison to the wild-type sequence (11) from EC-susceptible strain SC5314 (NCBI accession XM_716336) identified a single nucleotide change predicted to alter the protein sequence: T1922A (relative to the start codon), which changes residue 641 from F to Y. The sequence chromatogram clearly indicated only one base at the mutated position (data not shown); thus, the mutation was homozygous. The mutation was confirmed by sequencing independently amplified DNA.
FIG. 2.
Alignment of GS sequences corresponding to amino acids 641 to 648 of C. albicans Gsc1. BLASTP searches of the respective genome databases were used to align full-length Gsc1 with its C. albicans homologs Gsl2 and Gsl1 (11) and its C. glabrata and S. cerevisiae homologs Fks1, Fks2, and Fks3. Amino acids that are conserved relative to Gsc1 are represented as dots; amino acids whose mutations have been associated with RES are underlined and are shown below the relevant GS. TBLASTN searches of the NCBI (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism = fungi) and Broad Institute Fungal Genome Initiative (www.broad.mit.edu/annotation/fgi) databases were used to identify GS homologs from other species whose sequences predict RES based on alignment with the C. albicans, C. glabrata, and S. cerevisiae sequences and their RES mutants. To amplify and sequence the relevant GS regions in C. albicans 20464 or C. glabrata 20409, the following primer pairs were used: GSC1F (5′-TCATTGCTGTGGCCACTTTAG, located 1,718 bp from the start codon) and GSC1R (5′-TAGAATGAACGACCAATGGAGA, 2,135 bp); FKS1F (5′-GTTGCAGTCGCTACATTGCTA, 1,627 bp) and FKS1R (5′-TAGCGTTCCAGACTTGGGAA, 2,227 bp); FKS2F (5′-GGCCACTGTTTTATTCTTCTCG, 1,782 bp) and FKS2R (5′-GTAAATGTTCTCTGTACATGGA, 2,359 bp); and FKS3F (5′-TGGAGCCCAGCACTTAACAA, 1,389 bp) and FKS3R (5′-GTCCATCTCGGATGTTGCTA, 2,052 bp). For the amplification and sequencing of FKS2 from C. glabrata 2001-16 and 200989-16 transformants, FKS2F2 (5′-GTTCAATCAAAGGCAGGCCA, 1,747 bp) was used in place of FKS2F. Template DNA preparation and PCR amplification were performed as described previously (4). Following purification, PCR products were directly sequenced with one or both of the primers used to generate the product.
The BLAST analysis of the C. glabrata genome database (cbi.labri.fr/Genolevures/elt/CAGL) (3) revealed three GS homologs, and synteny with S. cerevisiae identified these as FKS1 (CAGL0G01034g), FKS2 (CAGL0K04037g), and FKS3 (CAGL0M13827g). The amplification and sequencing of the DNA regions of C. glabrata 20409 that were homologous to the GSC1 mutational hotspot identified no changes in FKS1 or FKS3 that would alter the predicted protein sequence, but in FKS2, a T1975G mutation was detected that would alter amino acid 659 from F to V. This mutation was confirmed in an independent PCR product. Since C. glabrata is haploid, no mixed bases were expected or observed.
Sequence alignment (Fig. 2) revealed that the C. albicans 20464 and C. glabrata 20409 mutations involve the analogous residue. This residue has not previously been implicated in C. albicans RES; however, its alteration has been associated with RES in an S. cerevisiae laboratory mutant (Fig. 2) (2).
Confirmation of the role in RES of the C. glabrata FKS2 mutation.
The EC-susceptible parents of C. glabrata 20409 and C. albicans 20464 were not available. We therefore pursued an alternative strategy to test the role of the identified mutations in RES. The strain 20409 DNA region that was sequenced was amplified again and transformed into EC-susceptible strain ATCC 2001, with selection on YPD agar containing 0.2 μg/ml caspofungin. Colonies were then screened for growth on 2 μg/ml caspofungin, and one clone (2001-16) showing strong growth was used to prepare DNA. The relevant FKS2 region was amplified using a forward primer located upstream of the original forward primer (see the legend for Fig. 2), which ensured that the chromosomal gene was amplified. Sequencing confirmed that clone 2001-16 incorporated the T1975G and hence the F659V mutation (Fig. 2). In broth microdilution assays, 2001-16 exhibited a caspofungin MIC twofold above that of its parent at 24 h which increased to four- to eightfold with longer incubation. This confirmation approach was independently repeated with a different C. glabrata strain (ATCC 200989), and the same result was obtained.
A similar attempt was made to transform EC-susceptible C. albicans (lab strain SC5314 and clinical isolate LL) with a GSC1 PCR product from mutant 20464, followed by selection on caspofungin-containing medium. This was not successful, presumably because a heterozygous GSC1 F641Y mutation was not sufficient to confer RES.
Analysis of fungal genome databases for F641 “mutation.”
The clinical usefulness of ECs is seriously compromised by the intrinsic RES exhibited by certain fungi. This could potentially be based on their GS sequences, although no examples of this have been reported to date. We searched available fungal genome sequences (see the legend to Fig. 2) for GS homologs and inspected the alignments for F641 alterations. The phytopathogenic molds Fusarium graminearum, Fusarium verticillioides, and Magnaporthe grisea, along with bread mold Neurospora crassa, have single GS homologs with Y641 equivalents (Fig. 2); i.e., the same residue found in C. albicans RES isolate 20464. This predicts that these molds should exhibit RES, which has indeed been reported for clinical isolates of related Fusarium species (5, 18). We tested caspofungin susceptibility of the sequenced N. crassa strain (74-OR23-1VA; Fungal Genetics Stock Center; no. 2489) and observed that it was susceptible (MIC, 0.03 μg/ml) but exhibited trailing growth (ca. 20% of control) at 24 h up to the highest concentration tested (2 μg/ml). Aspergillus nidulans (ATCC 10074) tested in parallel was similarly susceptible but exhibited no trailing growth at that time point.
This analysis also revealed that one of three GS genes in Candida (Pichia) guilliermondii has the equivalent of Y641 (Fig. 2). This gene is syntenic (as evidenced by the adjacent PWP2) with GSL2 of C. albicans. This Y641 may explain the intrinsic RES characteristic of C. guilliermondii (5, 16). In YPD medium, we observed a caspofungin MIC of 0.5 μg/ml for the sequenced C. guilliermondii strain ATCC 6260, which was about 32-fold above that obtained for representative C. albicans strains.
Conclusion.
These studies implicate the mutation of F641 in C. albicans Gsc1 or the equivalent residue of C. glabrata Fks2 as a potential basis for RES. Sequence alignments further suggest that substitutions involving this residue contribute to the intrinsic RES of molds such as Fusarium species and the yeast C. guilliermondii. The RES exhibited by yeast with F641 substitutions (MICs, 0.5 to 8 μg/ml) appears to be significantly less than that reported by Park et al. (14) for mutants with S645 mutations (MICs, ≥32 μg/ml); this probably explains why mutants with F641 mutations were not identified by those investigators. Our studies were limited by the lack of clinical data for the patients from which these RES strains were isolated. Nevertheless, these data add to the growing body of evidence that specific GS mutations confer acquired RES on susceptible fungi, while analogous substitutions may confer intrinsic resistance on others. Ultimately, these structure activity data should prove useful in modeling EC interaction with its GS target.
Acknowledgments
This work was supported by NIAID grants AI054289 (to S.K.) and AI47718 (to T.E.).
REFERENCES
- 1.Denning, D. W. 2003. Echinocandin antifungal drugs. Lancet 362:1142-1151. [DOI] [PubMed] [Google Scholar]
- 2.Douglas, C. M. 2001. Fungal beta(1,3)-D glucan synthesis. Med. Mycol. 39(Suppl. 1):55-66. [DOI] [PubMed] [Google Scholar]
- 3.Dujon, B, D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, et al. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 4.Edlind, T. D., K. W. Henry, J.-P. Vermitsky, M. P. Edlind, S. Raj, and S. K. Katiyar. 2005. Promoter-dependent disruption of genes: simple, rapid, and specific PCR-based method with application to three different yeast. Curr. Gen. 48:117-125. [DOI] [PubMed] [Google Scholar]
- 5.Espinel-Ingroff, A. 1998. Comparison of in vitro activities of the new triazole SCH56592 and the echinocandins MK-0991 (L-743872) and LY303366 against opportunistic filamentous and dimorphic fungi and yeasts. J. Clin. Microbiol. 36:2950-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez, S., J. L. Lopez-Ribot, L. K. Najvar, D. I. McCarthy, R. Bocanegra, and J. R. Graybill. 2004. Caspofungin resistance in Candida albicans: correlating clinical outcome with laboratory susceptibility testing of three isogenic isolates serially obtained from a patient with progressive Candida esophagitis. Antimicrob. Agents Chemother. 48:1382-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogh-Madsen, M., M. C. Arendrup, L. Heslet, and J. D. Knudsen. 2006. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 42:938-944. [DOI] [PubMed] [Google Scholar]
- 8.Kurtz, M. B., G. Abruzzo, A. Flattery, K. Bartizal, J. A. Marrinan, W. Li, J. Milligan, K. Nollstadt, and C. M. Douglas. 1996. Characterization of echinocandin-resistant mutants of Candida albicans: genetic, biochemical, and virulence studies. Infect. Immun. 64:3244-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtz, M. B., and J. H. Rex. 2001. Glucan synthase inhibitors as antifungal agents. Adv. Protein Chem. 56:423-475. [DOI] [PubMed] [Google Scholar]
- 10.Laverdiere, M., R. G. Lalonde, J. G. Baril, D. C. Sheppard, S. Park, and D. S. Perlin. 2006. Progressive loss of echinocandin activity following prolonged use for treatment of Candida albicans oesophagitis. J. Antimicrob. Chemother. 57:705-708. [DOI] [PubMed] [Google Scholar]
- 11.Mio, T., M. Adachi-Shimizu, Y. Tachibana, H. Tabuchi, S. B. Inoue, T. Yabe, T. Yamada-Okabe, M. Arisawa, T. Watanabe, and H. Yamada-Okabe. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol. 179:4096-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moudgal, V., T. Little, D. Boikov, and J. A. Vazquez. 2005. Multiechinocandin- and multiazole-resistant Candida parapsilosis isolates serially obtained during therapy for prosthetic valve endocarditis. Antimicrob. Agents Chemother. 49:767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohyama, T., S. Miyakoshi, and F. Isono. 2004. FKS1 mutations responsible for selective resistance of Saccharomyces cerevisiae to the novel 1,3-beta-glucan synthase inhibitor Arborcandin C. Antimicrob. Agents Chemother. 48:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, S., R. Kelly, J. N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier, R., I. Alarie, R. Lagace, and T. J. Walsh. 2005. Emergence of disseminated candidiasis caused by Candida krusei during treatment with caspofungin: case report and review of literature. Med. Mycol. 43:559-564. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., L. Boyken, R. J. Hollis, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2006. In vitro susceptibilities of Candida spp. to caspofungin: four years of global surveillance. J. Clin. Microbiol. 44:760-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller, M. A., R. N. Jones, S. A. Messer, M. B. Edmond, and R. P. Wenzel. 1998. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn. Microbiol. Infect. Dis. 30:121-129. [DOI] [PubMed] [Google Scholar]
- 18.Pfaller, M. A., F. Marco, S. A. Messer, and R. N. Jones. 1998. In vitro activity of two echinocandin derivatives, LY303366 and MK-0991 (L-743,792), against clinical isolates of Aspergillus, Fusarium, Rhizopus, and other filamentous fungi. Diagn. Microbiol. Infect. Dis. 30:251-255. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2003. Caspofungin activity against clinical isolates of fluconazole-resistant Candida. J. Clin. Microbiol. 41:5729-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safdar, A., V. Chaturvedi, B. S. Koll, D. H. Larone, D. S. Perlin, and D. Armstrong. 2002. Prospective, multicenter surveillance study of Candida glabrata: fluconazole and itraconazole susceptibility profiles in bloodstream, invasive, and colonizing strains and differences between isolates from three urban teaching hospitals in New York City (Candida Susceptibility Trends Study, 1998 to 1999). Antimicrob. Agents Chemother. 46:3268-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, W. L., and T. D. Edlind. 2002. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upreguluation. Antimicrob. Agents Chemother. 46:3532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]