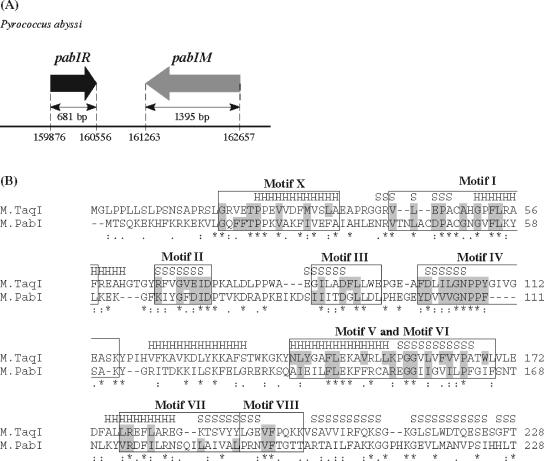
FIG. 1.
Organization of the PabI locus and comparative analysis of the N-terminal region of M.PabI. (A) PabI locus. Arrows represent coding regions of pabIR (PAB0105) and pabIM (PAB2246) in the genome sequence of P. abyssi (http://www.genoscope.cns.fr/Pab/). The length of each open reading frame is indicated in base pairs. The restriction gene pabIR and the putative modification gene pabIM are located in a tail-to-tail orientation, as is the case for many RM gene pairs (23). (B) Amino acid sequence alignment of M.PabI and M.TaqI (aligned usingClustal W version 1.82 [http://www.ebi.ac.uk/clustalw/index.html]). Symbols denoting the degree of conservation are indicated beneath the alignment: asterisks, double dots, and single dots represent identical, conserved, and semiconserved amino acids, respectively. Structural elements of α-helices (H) and β-strands (S), which were revealed by X-ray crystallographic analyses for M.TaqI (26), are indicated above the M.TaqI sequences. Assignments of nine conserved motifs as amino-methyltransferases (boxed areas) and consensus amino acids of group γ (shaded areas) are based on the structure-guided analysis by Malone et al. (27).