Abstract
Aromatic compounds represent an important source of energy for soil-dwelling organisms. The β-ketoadipate pathway is a key metabolic pathway involved in the catabolism of the aromatic compounds protocatechuate and catechol, and here we show through enzymatic analysis and mutant analysis that genes required for growth and catabolism of protocatechuate in the soil-dwelling bacterium Sinorhizobium meliloti are organized on the pSymB megaplasmid in two transcriptional units designated pcaDCHGB and pcaIJF. The pcaD promoter was mapped by primer extension, and expression from this promoter is demonstrated to be regulated by the LysR-type protein PcaQ. β-Ketoadipate succinyl-coenzyme A (CoA) transferase activity in S. meliloti was shown to be encoded by SMb20587 and SMb20588, and these genes have been renamed pcaI and pcaJ, respectively. These genes are organized in an operon with a putative β-ketoadipyl-CoA thiolase gene (pcaF), and expression of the pcaIJF operon is shown to be regulated by an IclR-type transcriptional regulator, SMb20586, which we have named pcaR. We show that pcaR transcription is negatively autoregulated and that PcaR is a positive regulator of pcaIJF expression and is required for growth of S. meliloti on protocatechuate as the carbon source. The characterization of the protocatechuate catabolic pathway in S. meliloti offers an opportunity for comparison with related species, including Agrobacterium tumefaciens. Differences observed between S. meliloti and A. tumefaciens pcaIJ offer the first evidence of pca genes that may have been acquired after speciation in these closely related species.
Aromatic acids constitute an important source of carbon and energy for soil-dwelling microorganisms and accumulate primarily as the result of the degradation of plant-derived molecules, including lignin. Many aromatic compounds may be converted to one of two common intermediates, protocatechuate or catechol, which are metabolized to tricarboxylic acid intermediates via the β-ketoadipate pathway (Fig. 1A) (18). In Agrobacterium tumefaciens, genes encoding enzymes involved in protocatechuate catabolism are organized into two distinct operons (36). Expression of the pcaDCHGB operon is induced by pathway metabolites β-carboxy-cis,cis-muconate and γ-carboxymuconolactone via the LysR-type transcriptional regulator protein PcaQ (35, 37). Genes in this operon are involved in the conversion of protocatechuate to the pathway intermediate β-ketoadipate. Expression of the pcaIJF operon is induced in the presence of β-ketoadipate (36), and these genes mediate the conversion of β-ketoadipate to the end products succinate and acetyl-coenzyme A (CoA). The transcriptional regulator involved in modulating expression of the pcaIJF operon in A. tumefaciens is an adjacent IclR-type regulator encoded by pcaR (39).
FIG. 1.
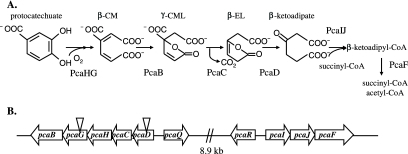
Protocatechuate catabolism in S. meliloti. (A) The protocatechuate branch of the β-ketoadipate pathway is involved in the catabolism of protocatechuate to intermediates which are funneled into the tricarboxylic acid cycle. β-CM, β-carboxy-cis,cis-muconate; γ-CML, γ-carboxymuconolactone; β-EL, β-ketoadipate enol-lactone. (B) Schematic depiction of the pca genes on the pSymB megaplasmid. Inverted triangles indicate the locations of transposon insertions in S. meliloti strains RmG879 (left triangle) and RmG867 (right triangle).
The gram-negative bacterium Sinorhizobium meliloti forms a symbiotic relationship with alfalfa through the establishment of root nodules. The β-ketoadipate pathway is present in many members of Rhizobiaceae examined to date, emphasizing the importance of aromatic acid catabolism in this family (41, 42). The publication of the S. meliloti genome has facilitated the identification and characterization of many metabolic pathways (10), and here we report the characterization of the protocatechuate branch of the β-ketoadipate pathway in S. meliloti. Except for pcaIJ orthologues, we demonstrate that the pca genes are organized, function, and are regulated in S. meliloti in a manner similar to that previously established for A. tumefaciens. Unexpectedly, the S. meliloti genes SMb20587 and SMb20588 were found to encode proteins with low sequence similarity to the two protein subunits of β-ketoadipate succinyl-CoA transferase (PcaI and PcaJ) in A. tumefaciens, Acinetobacter baylyi strain ADP1, and Pseudomonas putida. Through overexpression of SMb20587 and SMb20588 in S. meliloti followed by purification of β-ketoadipate succinyl-CoA transferase activity, these two genes are demonstrated to encode β-ketoadipate succinyl-CoA transferase activity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used throughout this study are described in Table 1. Escherichia coli was grown at 37°C in LB broth. Sinorhizobium meliloti was grown at 30°C in M9 minimal medium (Difco) or LB broth supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBmc). M9 minimal medium was supplemented with 1.0 mM MgSO4, 0.25 mM CaCl2, 1 μg/ml d-biotin, and 10 ng/ml CoCl2. Unless otherwise specified, carbon sources were added to M9 minimal medium as follows: 0.5% (vol/vol) glycerol, 15 mM arabinose, 30 mM adipate (Sigma-Aldrich), or 5 mM protocatechuate (Sigma-Aldrich). For S. meliloti, antibiotics were used at the following concentrations (in μg/ml): streptomycin, 200; neomycin, 200; gentamicin, 60; spectinomycin, 200; tetracycline, 5.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm1021 | Smr derivative of wild-type strain SU47 | 28 |
| Rm5004 | Rm1021 recA::Tn5 Smr Nmr | Strain collection |
| RmG212 | Rm1021 lac Smr | Strain collection |
| RmG867 | Rm1021 pcaD::Tn5 Smr Nmr | This study |
| RmG879 | Rm1021 pcaG::Tn5 Smr Nmr | This study |
| RmK927 | Rm5004 (pTH1459) Smr Tcr | This study |
| RmK948 | RmG212 pcaG::Tn5 Smr Nmr | This study |
| RmK1014 | Rm1021 pcaR::Ω Smr Spr | This study |
| RmK1015 | Rm1021 pcaF::gusA Smr Nmr | This study |
| RmK1016 | Rm1021 pcaF::gusA pcaR::Ω Smr Nmr Spr | This study |
| RmP134 | RmG212 pcaQ::Ω Smr Gmr | This study |
| RmP135 | RmG212 (pTH468) Smr Tcr | This study |
| RmP136 | RmP134 (pTH468) Smr Gmr Tcr | This study |
| RmP892 | Rm1021 (pTH1335) Smr Tcr | This study |
| RmP893 | RmK1014 (pTH1335) Smr Tcr | This study |
| RmP894 | RmK948 (pTH468) Smr Nmr Tcr | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) supE44 λ−thi-1 gyrA96 relA1 | Bethesda Research Laboratories, Inc. |
| BL21 | E. coli B F−ompT hsdSB(rB− mB−) dcm Tetrgal λ(DE3) endA Hte [argU proL Camr] | Stratagene |
| Plasmids | ||
| pLAFR1 | IncP cosmid cloning vector; Tcr | 12 |
| pFus1 | Broad-host-range gusA transcriptional reporter plasmid; Tcr | 45 |
| pMP220 | Broad-host-range lacZ transcriptional reporter plasmid; Tcr | 48 |
| pHP45Ω | pBR322 derivative with Ω element; Apr Spr Smr | 43 |
| pHP45Ωaac | pBR322 derivative with Ω element; Apr Gmr | 3 |
| pRK602 | pRK600Ω::Tn5, suicide vector used in transposon mutagenesis of Rm1021; Cmr Nmr | 9 |
| pJQ200 uc1 | Suicide vector with sacB to select for plasmid excision; Gmr | 44 |
| pVO155 | Suicide vector with promoterless gusA; Apr Nmr | 30 |
| pTH178 | pLAFR1 derivative complementing Pca− phenotype of RmG867 and RmG879; Tcr | This study |
| pTH468 | 434-bp EcoRI-XbaI PCR product encompassing pcaD-pcaQ intergenic region in pMP220 (pcaD::lacZ); Tcr | This study |
| pTH1227 | Broad-host-range derivative of pFus1 with Ptac promoter inserted upstream of gusA; Tcr | J. Cheng and T. M. Finan, unpublished data |
| pTH1335 | 153-bp EcoRI-PstI PCR product encompassing pcaR-pcaI intergenic region in pFus1 (pcaR::gusA); Tcr | This study |
| pTH1338 | 963-bp blunt-ended PCR product encompassing SacII site within pcaR, cloned into pUC119 via SmaI; Apr | This study |
| pTH1340 | ΩSm/Spr from pHP45Ω into pTH1338 via SacII; Apr Smr Spr | This study |
| pTH1351 | pcaR::Ω from pTH1340 into pJQ200 uc1 via NotI; Smr Spr Gmr | This study |
| pTH1360 | pVO155 derivative with gusA from pFus1; Apr Nmr | R. Zaheer and T. M. Finan, unpublished data |
| pTH1459 | 1,701-bp EcoRI-PstI PCR product encompassing pcaIJ in expression vector pTH1227; Tcr | This study |
| pTH1559 | 431-bp SpeI-XbaI PCR product encompassing 3′ end of pcaF into pTH1360 (pcaF::gusA); Apr | This study |
| pTH1577 | 893-bp NotI PCR product encompassing PstI site within pcaQ cloned into pJQ200 uc1; Gmr | This study |
| pTH1592 | ΩGmr from pHP45Ω into pTH1577 via PstI site; Gmr | This study |
| pTH1882 | pcaQ::Ω from pTH1592 into pTH1883 via NotI; Apr Nmr Gmr | This study |
| pTH1883 | pTH1360 with gusA removed via NotI digest; Apr Nmr | This study |
Transposon mutagenesis.
Tn5 mutagenesis of S. meliloti was performed by mating the suicide plasmid pRK602 into wild-type derivative strain Rm1021. Neomycin-resistant colonies were patched onto M9 minimal media with protocatechuate or glucose as the sole carbon source. Mutants unable to grow on protocatechuate (Pca−) were examined for ability to grow with succinate as a sole carbon and energy source to eliminate mutants deficient in succinate metabolism.
The pLAFR1 clone bank of S. meliloti Rm1021 DNA (12) was screened to isolate clones capable of complementing Pca− strains. Spot matings were performed with Pca− mutants, using the clone bank, and strains carrying the complementing cosmids were selected on M9 minimal media with protocatechuate as a sole carbon source. DNA sequencing was provided by Mobix (McMaster University, Hamilton, Ontario, Canada).
Rothera test.
An overnight LBmc culture was centrifuged and washed with M9 minimal medium. Cells were subcultured into M9 minimal medium supplemented with 0.1% arabinose and 5 mM protocatechuate for overnight incubation at 30°C. Cells were centrifuged and resuspended in 0.02 M Tris-HCl, pH 8.0, to an optical density (OD) of 1.0. Toluene (0.5 ml) was added to 2 ml resuspended cells, which was incubated at 30°C with shaking for 1 h. (NH4)2SO4 (1 g) was added, and the mixture was vortexed. One drop of a fresh aqueous sodium nitroprusside (1%) solution was added, followed by the addition of 1 drop of concentrated NH3 (29%), and the mixture was vortexed. Development of a purple color within 5 min following the addition of NH3 was considered a positive test for the presence of β-ketoadipate (20).
Protocatechuate 3,4-dioxygenase activity assays.
Overnight cultures grown in LBmc were washed and subcultured into M9 minimal medium supplemented with 0.1% arabinose and 5 mM protocatechuate. Tetracycline was included in the growth medium for strains carrying pTH178. Upon harvest, cells were centrifuged, washed, and resuspended into 4 ml buffer (20 mM Tris-HCl, 1 mM MgCl2, pH 7.8) per gram cells. Aliquots of cells were frozen at −80°C until used in the assay.
Prior to use, 10 μl of 0.1 M dithiothreitol was added (per ml) as the aliquots thawed on ice. Cells were disrupted via sonication, and extracts were centrifuged to remove intact cells and cellular debris. The dioxygenase assay was performed as previously described (8), with the following modifications. The temperature of the assay was maintained at 30°C, and the reaction was monitored by following the reduction in absorbance at 293 nm using a Contron Uvikon 930 double-beam spectrophotometer. Protein concentrations were determined using Bio-Rad protein assay reagent, with bovine serum albumin as the standard.
Construction of an S. meliloti pcaQ::Ω strain.
An Ω cassette encoding gentamicin resistance from pHP45Ωaac (3) was introduced into a PstI site located 35 bp downstream of the predicted pcaQ translational start site as follows. An 893-bp fragment centered upon the PstI site was PCR amplified using S. meliloti Rm1021 genomic DNA as a template. This fragment was cloned into the suicide vector pJQ200 uc1 (44) via NotI to create plasmid pTH1577. The Ω cassette was PCR amplified and cloned into pTH1577 via PstI, yielding pTH1592. The NotI fragment, encompassing pcaQ::Ω, was subcloned from pTH1592 into a derivative of pVO155 (30) to create pTH1882. This pVO155 derivative (pTH1883) lacks the gusA reporter gene present in the parental vector and was selected for use because it is unable to replicate in S. meliloti and carries a gene specifying neomycin resistance. pTH1882 was mated into S. meliloti lac strain RmG212, and recombinants were selected for by plating onto LB agar supplemented with streptomycin plus gentamicin. Colonies were patched onto LB agar plus neomycin to identify recombinants in which the suicide vector had recombined out of the genome, leaving the Ω cassette behind. Southern hybridization was performed on DNA extracted from Gmr Nms recombinants to confirm the location of the antibiotic cassette and absence of the suicide vector. In this case, an EcoRV digest of genomic DNA isolated from putative RmG212 pcaQ::Ω mutants and RmG212 was hybridized with a labeled (random primed DNA labeling kit; Roche) probe encompassing the pcaQ PstI site. A shift corresponding to an ∼2-kb increase (compared to the wild type) was noted in the putative pcaQ mutants, consistent with the incorporation of the 1.8-kb Gmr cassette into the PstI site and the subsequent excision of the integrating vector. As well, hybridization of the EcoRV genomic DNA digest with labeled probe corresponding to the (Gmr) Ω cassette indicated the presence of the antibiotic cassette within the shifted bands observed in the putative mutants. The Ω probe failed to hybridize with the EcoRV-digested RmG212 genomic DNA.
Construction of an S. meliloti pcaR::Ω strain.
An Ω cassette encoding streptomycin/spectinomycin resistance from pHP45Ω (43) was introduced into a SacII site located 76 bp downstream of the predicted pcaR translational start site. A 963-bp fragment encompassing the SacII site within pcaR was PCR amplified using S. meliloti Rm1021 DNA as a template and Vent DNA polymerase (New England Biolabs). This blunt-ended fragment was cloned into pUC119 at a SmaI site to create plasmid pTH1338. The Ω cassette was PCR amplified and cloned into the SacII site in pTH1338 to produce pTH1340. A NotI fragment, encompassing pcaR::Ω, was subcloned from pTH1340 into the suicide vector pJQ200 to yield pTH1351. This plasmid was mated into S. meliloti Rm1021 and recombinants were selected for by plating onto LB agar supplemented with streptomycin plus gentamicin. A single Gmr colony was inoculated into LBmc and grown in the absence of antibiotic selection. The overnight culture was plated onto LB agar supplemented with 5% sucrose and spectinomycin. Spr colonies were patched onto LB agar plus gentamicin to confirm excision of the suicide plasmid. Southern hybridization was performed on DNA extracted from Spr Gms sucrose-resistant colonies to confirm the location of the Ω cassette and verify the loss of the integrating plasmid. Briefly, separate XhoI and SalI digests of genomic DNA isolated from putative Rm1021 pcaR::Ω mutants and Rm1021 were hybridized with a labeled (random primed DNA labeling kit; Roche) probe encompassing the pcaR SacII site. In each case, a shift corresponding to an ∼2-kb increase (compared to the wild type) was noted in the putative pcaR mutants, consistent with the incorporation of the 2.1-kb antibiotic cassette into the SacII site and the excision of the integrating vector. As well, hybridization of separate XhoI and SalI genomic DNA digests with labeled probe corresponding to the (Smr Spr) Ω cassette indicated the presence of the antibiotic cassette within the shifted bands observed in the putative mutants. The Ω probe failed to hybridize with either SalI- or XhoI-digested Rm1021 genomic DNA.
Construction of an S. meliloti pcaF::gusA strain.
A transcriptional fusion between the annotated pcaF gene on the pSymB megaplasmid and a promoterless gusA gene was created in S. meliloti wild-type derivative Rm1021 and PcaR− strain RmK1014. The fusion was designed such that pcaF was not disrupted, and this was verified by growth with protocatechuate as a sole carbon source. A 431-bp fragment spanning the 3′ end of pcaF was PCR amplified and cloned into the pVO155 (30) derivative pTH1360 to create pTH1559. In pTH1360, the original gusA reporter gene present in pVO155 has been replaced by the gusA gene present in pFus1, which has a superior ribosome binding site and is expressed more efficiently than its pVO155 counterpart (R. Zaheer and T. M. Finan, unpublished data). pTH1559 was mated into Rm1021 and RmK1014, and recombination of the vector into the S. meliloti genome was selected by plating cells onto LB agar supplemented with streptomycin plus neomycin.
Purification of β-ketoadipate succinyl-CoA transferase activity in S. meliloti.
β-Ketoadipate succinyl-CoA transferase was purified according to the method of Kaschabek et al. (24), with some modifications. An overnight culture of RmK927 was subcultured into 4 liters LBmc supplemented with tetracycline, and cells were grown with shaking at 30°C. Expression of genes SMb20587 and SMb20588 was induced at an OD of 0.3 to 0.4 with the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 5 mM protocatechuate. After 4 h of induction, cells were harvested in the late exponential growth phase (OD of 1.2 to 1.4). The pellet was resuspended in 30 ml buffer (100 mM Tris-HCl, 0.5 mM dithiothreitol, pH 7.0) and then lysed via five passages through a French pressure cell at 110 MPa. The cell extract was cleared by centrifugation at 100,000 × g for 60 min. Solid (NH4)2SO4 was added to give 75% saturation, and the precipitate was collected by centrifugation at 8,000 × g for 20 min. The pellet was redissolved in buffer B1 [50 mM Tris-HCl, 1 M (NH4)2SO4, 1 mM EDTA, pH 7.0], and the extract was cleared by centrifugation. The supernatant was loaded onto a phenyl Sepharose CL-4B (Amersham Biosciences) column preequilibrated with buffer B1, and protein was eluted in a linear gradient of (NH4)2SO4 from 1 to 0 M at a flow rate of 0.3 ml/min. Nine 2-ml fractions with β-ketoadipate succinyl-CoA transferase activity were pooled and dialyzed into buffer A1 (50 mM Tris-HCl, 0.5 mM EDTA, pH 7.0).
The dialyzed fractions were loaded onto a Source 30Q (Amersham Biosciences) column preequilibrated with buffer A1, and protein was eluted in a linear gradient of NaCl from 0 to 1 M at a flow rate of 0.3 ml/min. Twelve 0.5-ml fractions with β-ketoadipate succinyl-CoA transferase activity were pooled and dialyzed into 5 mM potassium phosphate, pH 7.0.
The fractions were loaded onto a CHT ceramic hydroxyapatite (Bio-Rad) column preequilibrated with 5 mM potassium phosphate, pH 7.0. Elution occurred in a linear gradient of potassium phosphate from 10 to 400 mM at a flow rate of 0.3 ml/min. Twelve 1-ml fractions with β-ketoadipate succinyl-CoA transferase activity were collected and pooled. An attempt was made to concentrate the enzyme within the pooled extract by using Nanosep microconcentrators; however, the majority of the purified enzyme was lost at this step, as revealed by subsequent β-ketoadipate succinyl-CoA transferase assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by staining with Coomassie brilliant blue.
β-Ketoadipate succinyl-CoA transferase assays.
β-Ketoadipate succinyl-CoA transferase assays were performed as previously described (54), with the following modifications. Briefly, the enzyme assays were performed with UV-Star (flat-bottomed) 96-well microtiter plates (Greiner Bio-One) and a Safire microplate reader (Tecan). To start the reaction, protein samples were added to a buffered reaction mixture (200 mM Tris-HCl, 40 mM MgCl2, 10 mM β-ketoadipate [Sigma-Aldrich], 0.4 mM succinyl-CoA, pH 8.0) to a final volume of 0.2 ml (path length, 0.52 cm). The formation of β-ketoadipyl-CoA:Mg2+ was monitored at 305 nm over a temperature range of 22 to 23°C. An extinction coefficient of 16,300 M−1 cm−1 was used to calculate the formation of the β-ketoadipyl-CoA:Mg2+ complex (24). One unit of activity is defined as the amount of enzyme required to convert 1 μmol of substrate to product in 1 min under the conditions of the assay.
Protein identification via mass spectrometry.
Mass spectrometry analyses were provided by the McMaster Regional Centre for Mass Spectrometry (McMaster University, Hamilton, Ontario, Canada). The following peptides were used in the identification of the proteins: with a molecular mass of 37.6 kDa, peptides R.NGNVLIEGIVGVQK.E, R.MTPDILYDQLIGVGAAR.G, and R.IMSLAEAVEENVR.D; with a molecular mass of 35.4 kDa, peptides R.NGNVLIEGIVGVQK.E and R.MTPDILYDQLIGVGAAR.G; and with a molecular mass of 28.5 kDa, peptides R.FANLNTTVVGPYDHPK.V, K.FAETVIETPAPTETELVVLR.D, and R.ITTGFLGGAQIDR.F.
Isolation of total RNA from S. meliloti.
Cultures were grown with shaking at 30°C in LBmc ± 5 mM protocatechuate to an OD of 0.6 to 0.7. Total RNA was isolated from S. meliloti Rm1021 by use of a hot phenol method as previously described (27).
Primer extension.
Primer extension reactions were performed using 50 μg of total S. meliloti RNA as previously described (27). The following primers were used for extension reactions: for the identification of the pcaD start site, 5′ GAAATCCGTGCCGAGCGAGTTGATGAAGAC 3′, and for the identification of the pcaI start site, 5′ GAGAGACATTATCCGCGCCATCG 3′ and 5′ CGTCTCTGACATTCTCCTCTACCG 3′. Sequencing reactions were performed using a Sequenase version 2.0 DNA sequencing kit (USB). The same primer was used in sequencing and primer extension reactions.
β-Galactosidase enzyme assays.
S. meliloti cultures were grown overnight at 30°C in LBmc and washed with 0.85% saline prior to subculture. Cells were subcultured into M9 minimal media supplemented with a carbon source as indicated and grown with shaking at 30°C for 4 h. Aliquots (50 to 200 μl) of cells were added directly to Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 40 mM, β-mercaptoethanol, pH 7.0) with 5% chloroform and 0.0025% SDS. The reaction was started with the addition of 200 μl 2-nitrophenyl β-d-galactopyranoside (ONPG) (4 mg/ml) and stopped upon addition of 500 μl 1 M Na2CO3. β-Galactosidase activities were calculated according to the method of Miller (29).
Enzyme assays were performed using derivatives of RmG212 (Rm1021 lac) to reduce background LacZ enzyme activity. RmK948 was created through the transduction of pcaG::Tn5 into RmG212, using RmG879 as a donor strain.
β-Glucuronidase enzyme assays.
Overnight LBmc cultures of S. meliloti were grown and washed with 0.85% saline. Cells were subcultured into M9 minimal media supplemented with a carbon source as indicated and grown with shaking at 30°C for 4 h. Cultures were centrifuged and resuspended into a buffer consisting of 50 mM sodium phosphate, 50 mM dithiothreitol, and 1 mM EDTA, pH 7.0. Enzyme assays were performed according to the method of Reeve et al. (45), with the exception that assays were performed at room temperature (20 to 22°C).
RESULTS AND DISCUSSION
Isolation and characterization of pca Rm1021 mutants.
To directly identify genes involved in protocatechuate catabolism in S. meliloti, we screened a transposon Tn5 insertion library for mutants able to grow with succinate but unable to grow with protocatechuate (Pca−) as a sole carbon source. The precise insertion sites of the transposon in the S. meliloti genome were determined by DNA sequencing (see Materials and Methods). Strains RmG867 and RmG879 were found to carry Tn5 within genes annotated to encode β-ketoadipate enol-lactone hydrolase (PcaD) and protocatechuate 3,4-dioxygenase alpha-subunit (PcaG), respectively. Analysis of the S. meliloti genome sequence suggests that pcaD and pcaG are organized in an operon with a γ-carboxymuconolactone decarboxylase (pcaC), protocatechuate 3,4-dioxygenase β-subunit (pcaH), and β-carboxy-cis,cis-muconate cycloisomerase (pcaB) (Fig. 1B) (10).
To determine whether β-ketoadipate accumulated from protocatechuate metabolism in either the pcaD or the pcaG mutant, we performed the Rothera test, which detects the presence of β-ketoadipate and thus indicates whether protocatechuate has been metabolized to this pathway intermediate. The wild-type strain Rm1021 exhibited a Rothera-positive phenotype, whereas the mutants RmG867(pcaD) and RmG879(pcaG) were Rothera negative, indicating that these strains failed to metabolize protocatechuate to β-ketoadipate (Fig. 1A). Protocatechuate 3,4-dioxygenase catalyzes the first step in the degradation of protocatechuate, the conversion of protocatechuate to β-carboxy-cis,cis-muconate (Fig. 1A) (31). Examination of S. meliloti extracts from cultures grown in minimal medium containing arabinose and protocatechuate revealed that protocatechuate 3,4-dioxygenase activity was readily detected in the parent strain, Rm1021, whereas no activity was detected in RmG867(pcaD) and RmG879(pcaG) (data not shown). Since the dioxygenase consists of protein subunits encoded by two genes (pcaHG), disruption of either of these genes (as in RmG879) would result in a corresponding loss of protocatechuate 3,4-dioxygenase activity. The enzyme β-ketoadipate enol-lactone hydrolase (PcaD) mediates the conversion of β-ketoadipate enol-lactone to β-ketoadipate (31), and a mutation in pcaD (as in RmG867) would block this step of the pathway, preventing the production of β-ketoadipate (Fig. 1A). Organization of the pcaDCHGB operon ensures that insertion of a transposon within pcaD would also disrupt expression of downstream pca genes, including pcaHG (Fig. 1B). The loss of protocatechuate 3,4-dioxygenase activity in RmG867(pcaD) presumably results from the polar nature of the mutation in this strain and supports the assumption that the genes in question are organized as a single transcriptional unit.
An S. meliloti Rm1021 pLAFR1 clone (12), pTH178, was isolated on the basis of its ability to complement the Pca− phenotype of the RmG867 and RmG879 mutants. The presence of the pTH178 plasmid in strains RmG867 and RmG879 restored a Rothera-positive phenotype and protocatechuate 3,4-dioxygenase activity to the RmG867 and RmG879 mutant strains (data not shown). Consistent with its ability to complement pcaD and pcaG mutant phenotypes, DNA sequencing of pTH178 revealed that this cosmid carries the pcaDCHGB region in its entirety.
Regulation of expression of the pcaDCHGB operon.
In S. meliloti, the pcaDCHGB operon is located adjacent to, and transcribed divergently from, a gene encoding a product with similarity to the A. tumefaciens LysR-type transcriptional regulator PcaQ. To examine the possible involvement of PcaQ in regulating pcaDCHGB expression, an S. meliloti interposon knockout mutant of pcaQ was constructed. The pcaQ mutant strain was unable to utilize protocatechuate as a sole carbon source; however, growth of the mutant strain was comparable to that of the wild type in media containing glucose or glycerol as the carbon source (data not shown).
To monitor expression from the pcaD promoter, the pcaD-pcaQ intergenic region was cloned into pMP220 (48), with the pcaD promoter in the same orientation as a promoterless lacZ reporter gene. The resulting replicating plasmid, pTH468, was conjugated into RmG212 (lac mutant Rm1021 derivative), RmP134 (pcaQ::Ω derivative of RmG212), and RmK948 (pcaG::Tn5 derivative of RmG212), a strain that is incapable of metabolizing protocatechuate to β-carboxy-cis,cis-muconate. Expression of pcaD (as measured by β-galactosidase activity) was examined following growth in minimal medium containing glycerol as a carbon source and in medium containing glycerol plus protocatechuate. In the wild-type background, expression of pcaD was induced greater than 10-fold in the presence of protocatechuate, while pcaD expression did not increase under similar growth conditions in RmP134(pcaQ) (Table 2). This suggests that a product encoded by pcaQ is required for induction of pcaD expression. The low level of expression of pcaD in both the uninduced wild-type and pcaQ mutant backgrounds suggests that PcaQ does not act as a repressor of pcaD transcription. Expression of pcaD was also not induced in the pcaG mutant background (RmK948), and this suggests that protocatechuate itself does not act as an inducing agent. Presumably, an intermediate in the β-ketoadipate pathway is required to induce pcaDCHGB. Given the similarity that exists in genetic organization and regulation of the β-ketoadipate pathways in S. meliloti and A. tumefaciens (38), it is likely that β-carboxy-cis,cis-muconate and γ-carboxymuconolactone serve as coinducers in the PcaQ-regulated expression of the pcaDCHGB operon in S. meliloti; however, this matter was not examined further.
TABLE 2.
Expression of pcaD-lacZ fusion in S. meliloti strains
| Strain | Relevant genotype | Growth condition | β-Galactosidase activitya (Miller units [SD]) |
|---|---|---|---|
| RmP135 | Rm1021 lac (pTH468) | Glycerol | 590 (18.5) |
| Glycerol plus PCAb | 8,210 (288.6) | ||
| RmP136 | Rm1021 lac pcaQ::Ω (pTH468) | Glycerol | 530 (39.6) |
| Glycerol plus PCA | 311 (8.6) | ||
| RmP894 | Rm1021 lac pcaG::Tn5 (pTH468) | Glycerol | 468 (36.3) |
| Glycerol plus PCA | 550 (20.2) |
Shown are averages of values obtained from three independent cultures grown overnight in LBmc and subcultured into M9 minimal medium with 0.5% glycerol ± 5 mM protocatechuate at 30°C for 4 h.
PCA, protocatechuate.
Identification of pcaDCHGB transcriptional start sites.
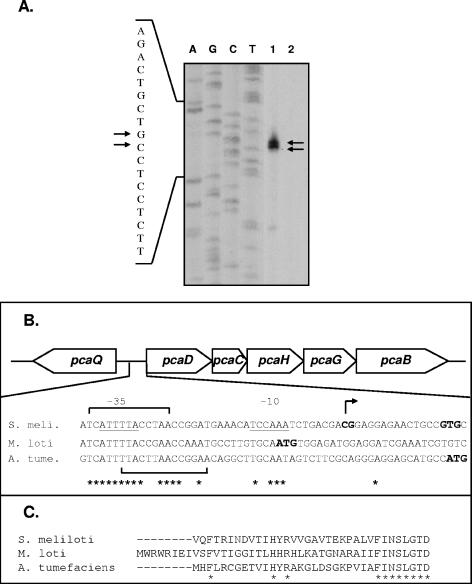
Primer extension analysis was performed on RNA isolated from S. meliloti strain Rm1021 grown in LBmc in the presence and absence of protocatechuate prior to RNA isolation. A 30-mer oligonucleotide complementary to the 5′ pcaD coding region was used to prime the extension reaction. Two major extension products were obtained only with template RNA isolated from cells grown in the presence of protocatechuate, indicating transcriptional start sites at G and C residues located 14 and 15 nucleotides upstream of the predicted pcaD translational start site, respectively (Fig. 2A).
FIG. 2.
Analysis of the pcaD promoter in S. meliloti. (A) Primer extension reactions were performed using RNA isolated from S. meliloti strain Rm1021 grown in the presence (lane 1) and absence (lane 2) of 5 mM protocatechuate. Sequencing reactions were performed with the same primer used in primer extension reactions, and results are shown to the left of the extension products. The arrows indicate the extension products (right) and identify the corresponding nucleotides (left). (B) Schematic depiction of the pcaQ-pcaDCHGB genes, including an alignment of the S. meliloti (S. meli.) pcaDp region with the sequence upstream of pcaD in M. loti and A. tumefaciens (A. tume.) (GenBank accession numbers NC_003078, NC_002678, and NC_003305, respectively). The two S. meliloti pcaD transcriptional start sites are identified in bold, and the inferred −10 and −35 regions are underlined. Nucleotides conserved among all three species are indicated by an asterisk, and predicted translational start sites are indicated (bold, enlarged font). Two putative PcaQ binding sites present in S. meliloti, A. tumefaciens, and M. loti are indicated by brackets. (C) Alignment of PcaD amino acid sequences (N terminus), as annotated in S. meliloti, M. loti, and A. tumefaciens genome sequences. Invariant residues are indicated by an asterisk. Sequences were aligned using CLUSTAL W (50).
The sequence upstream of the pcaD transcriptional start sites has AT-rich sequences centered at −10 and −35 hexanucleotide regions (Fig. 2B). Alignment of the sequences upstream of the pcaD genes from S. meliloti and A. tumefaciens with the region upstream of the annotated Mesorhizobium loti pcaD gene indicates that these regions are conserved among the three species (Fig. 2B). However, the M. loti sequence that corresponds to the inferred S. meliloti −10 hexanucleotide includes the annotated M. loti pcaD translational start site. Alignment of the PcaD amino acid sequences from S. meliloti, A. tumefaciens, and M. loti reveals that the M. loti protein consists of an additional 8 amino acids at the N terminus that are absent in the other two species (Fig. 2C). These results suggest that the correct start codon for pcaD in M. loti is located 24 bp downstream of the annotated site. Interestingly, this would mean that the M. loti gene employs a GTG translational start codon, as is predicted for pcaD in S. meliloti.
Examination of the pcaD promoter region also reveals potential PcaQ binding sites, several of which have also been conserved among S. meliloti, A. tumefaciens, and M. loti. As a member of the LysR family of transcriptional regulators, PcaQ may be expected to recognize and bind elements established upon a (T-N11-A) consensus binding motif (16, 47). Examination of the pcaD-pcaQ intergenic region from S. meliloti revealed seven T-N11-A motifs. Of these, two T-N-A-N9-A motifs span the −35 region in S. meliloti and are conserved in A. tumefaciens and M. loti (Fig. 2B). As pcaD and pcaQ are separated by a 94-bp intergenic region, it is possible that binding of PcaQ to a given site(s) may simultaneously exert positive and negative effects with respect to the expression of pcaD and pcaQ, respectively, as has been shown for the crgA-mdaB genes in Neisseria meningitidis (6, 21). PcaQ-mediated autoregulation has been described previously for A. tumefaciens (37), and we similarly have evidence of autoregulation in S. meliloti (data not shown). This work is being pursued using purified PcaQ to facilitate the identification of PcaQ binding sites.
Identification and purification of PcaIJ.
Genes encoding the β-ketoadipate succinyl-CoA transferase proteins (PcaIJ) have not been identified in the S. meliloti genome (13). Two genes (SMb20587 and SMb20588) are annotated as encoding subunits of a CoA transferase, and these are located approximately 10 kb from the pcaDCHGB operon. The SMb20587 and SMb20588 genes lie upstream of the pcaF gene, annotated to encode β-ketoadipyl-CoA thiolase. The low amino acid sequence similarity of SMb20587 and SMb20588 with other PcaIJ proteins prevented their annotation as PcaIJ orthologues.
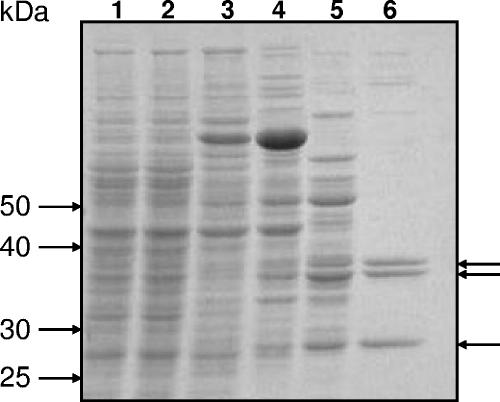
To establish whether SMb20587 and SMb20588 encode β-ketoadipate succinyl-CoA transferase activity, these genes were cloned into pTH1227, an IPTG-inducible expression vector carrying the tac promoter, to give pTH1459. S. meliloti (pTH1459) cells were induced with both protocatechuate and IPTG. Whole-cell lysate obtained from induced cultures exhibited β-ketoadipate succinyl-CoA transferase activity as detected spectrophotometrically by the increase in absorbance at 305 nm that accompanies formation of the β-ketoadipyl-CoA:Mg2+ complex (see Materials and Methods). This activity was sequentially purified to near homogeneity by use of a combination of ammonium sulfate precipitation and chromatography with columns containing phenyl Sepharose CL-4B, Source 30Q, and CHT ceramic hydroxyapatite (see Materials and Methods for details) (Fig. 3). SDS-PAGE analysis demonstrated the presence of a 28-kDa band that was consistent with the predicted size (27.7 kDa) of the β-subunit encoded by SMb20588. A second, higher-molecular-mass protein was present as a doublet of 36- and 38-kDa bands, which we thought could be an anomalously migrating species of the alpha-subunit (predicted size, 31.2 kDa) encoded by SMb20587. To positively identify these polypeptide species, all three bands were subjected to a trypsin digest and tandem mass spectrometry. The 28-kDa protein was confirmed as the β-subunit of a CoA transferase encoded by gene SMB20588. Both the 36- and 38-kDa proteins were identified as the alpha-subunits of a CoA transferase encoded by gene SMB20587. We have not explored why the alpha-subunit is expressed as a doublet, but it may be because expression was induced both from the pSymB megaplasmid (using protocatechuate as an inducer) and from an IPTG-inducible expression vector. Possibly, an alternative start codon was used in translation from the expression vector. Examination of the nucleotide sequence encoding the alpha-subunit revealed two potential in-frame start codons downstream of the annotated start site. Translation initiation from these alternative start sites would generate proteins differing by either 4 or 22 amino acids relative to the full-length protein. The difference in molecular mass between the doublet proteins as estimated from SDS-PAGE is ∼2 kDa (or approximately 20 amino acids), and this difference might therefore be explained by the use of two distinct translational start sites. In any case, polypeptides encoded by SMb20587 and SMb20588 are enriched to near homogeneity in a protein sample purified solely on the basis of β-ketoadipate succinyl-CoA transferase activity.
FIG. 3.
Purification of β-ketoadipate succinyl-CoA transferase in S. meliloti. S. meliloti Rm5004 carrying pTH1459 was grown in M9 minimal medium with 5 mM protocatechuate and 1 mM IPTG. Protein samples were subjected to SDS-PAGE in a 10% gel followed by staining with Coomassie brilliant blue. Lane 1, crude cell lysate derived from uninduced culture; lane 2, crude cell lysate obtained from cells induced by 1 mM IPTG and 5 mM protocatechuate; lane 3, ammonium sulfate precipitate of induced cell lysate; lane 4, pooled eluate from phenyl Sepharose column; lane 5, pooled eluate from Source 30Q (anion-exchange) column; lane 6, pooled eluate from hydroxyapatite column. Molecular masses are shown on the left. Arrows (right) indicate the proteins identified by mass spectrometry. The 36- and 38-kDa proteins were identified as the alpha-subunits of a CoA transferase encoded by gene SMb20587; the 28-kDa protein was identified as the β-subunit of a CoA transferase encoded by gene SMb20588.
The transfer of CoA to β-ketoadipate has been documented previously as resulting from the nonspecific activity of an adipate succinyl-CoA transferase (5, 19). To eliminate the possibility that an adipate succinyl-CoA transferase had been purified inadvertently, we performed enzyme assays with β-ketoadipate and with increasing concentrations of adipate to examine substrate specificity (Table 3). Addition of equimolar amounts of adipate did not result in a significant decrease in enzyme activity (7% decrease in activity); however, a fivefold-greater concentration of adipate (relative to β-ketoadipate) in a reaction reduced activity by 56%. These results indicate that although adipate might compete with β-ketoadipate as a substrate when present in a greater concentration, β-ketoadipate is the preferred substrate of this enzyme. Enzyme activity was also dependent upon the presence of β-ketoadipate, succinyl-CoA, and Mg2+, and omission of any one of these reagents from the reaction mixture abolished activity (<0.01 μmol/min/mg) (data not shown). We therefore concluded that SMb20587 and SMb20588 encode subunits of a β-ketoadipate succinyl-CoA transferase, and these genes were named pcaI and pcaJ, respectively.
TABLE 3.
β-Ketoadipate succinyl-CoA transferase activity in the presence of adipatea
| Assay condition | Ratio of adipate to β-ketoadipate | Transferase activity (mU/mg protein [SD]) | % Transferase activity (SD) |
|---|---|---|---|
| 0 mM adipate | 539 (12.6) | 100 (2.3) | |
| 2 mM adipate | 1:5 | 562 (24.4) | 104 (4.5) |
| 10 mM adipate | 1:1 | 503 (24.2) | 93 (4.5) |
| 50 mM adipate | 5:1 | 236 (24.0) | 44 (4.5) |
Enzyme assays were performed using purified enzyme. One unit of enzyme is the amount required to convert 1 μmol of β-ketoadipate to β-ketoadipyl-CoA in 1 min under assay conditions.
The β-ketoadipate pathway is composed of protocatechuate and catechol branches, and either pca or cat genes (or both) may be present within a given species. Accordingly, β-ketoadipate succinyl-CoA transferase activity may be encoded by either pca and/or cat genes (pcaIJ and catIJ, respectively), and these may share sequence similarity (15, 25). Amino acid sequence identity between the S. meliloti PcaIJ and that of A. baylyi (25) (PcaI, 21%; PcaJ, 20%), P. putida (33) (PcaI, 21%; PcaJ, 23%), Bradyrhizobium japonicum (23) (PcaI, 17%; PcaJ, 20%), and even A. tumefaciens (53) (PcaI, 19%; PcaJ, 21%) is limited. In contrast, the sequence identity that exists between the S. meliloti PcaIJ and that of M. loti (22) (PcaI, 63%; PcaJ, 71%) and Pseudomonas aeruginosa PAO1 (49) (PcaI, 70%; PcaJ, 60%) and Pseudomonas sp. strain B13 CatIJ (15) (CatI, 68%; CatJ, 60%) is extensive. Likewise, signature sequences typically present in PcaI and PcaJ of many species are absent or modified in their S. meliloti counterparts, a situation comparable to that previously described by Göbel et al. (15). For PcaI, an N-terminal glycine cluster ([DN]-[GN]-X[2]-[LIVMFA][3]-G-G-F-X[3]-G-X-P) (52) present in A. tumefaciens, B. japonicum, A. baylyi, and P. putida proteins has been modified in S. meliloti by the deletion of one glycine residue and the replacement of another with glutamic acid. This modification has been reported previously for CatI of Pseudomonas sp. strain B13 (15) and may also be observed to occur in P. aeruginosa and M. loti pcaIJ. For PcaJ, an N-terminal signature sequence ([LF]-[HQ]-S-E-N-G-[LIVF][2]-[GA]) (33) present in B. japonicum, A. baylyi, and P. putida is absent in S. meliloti. The E-S-G motif reported for CatJ of Pseudomonas sp. strain B13 (15) and present in P. aeruginosa, M. loti, and a glutaconate-CoA transferase of Acidaminococcus fermentans is also conserved in S. meliloti. Thus, the differences observed between β-ketoadipate succinyl-CoA transferases of one group of species (A. baylyi, P. putida, A. tumefaciens, and B. japonicum) and another (Pseudomonas sp. strain B13, P. aeruginosa, M. loti, and S. meliloti) are striking, especially since closely related organisms, such as A. tumefaciens and S. meliloti, carry different forms of the enzyme.
Identification of pcaI transcriptional start site.
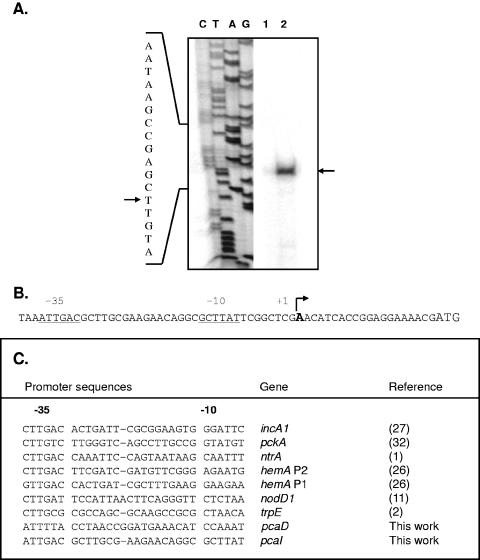
To facilitate our analysis of transcriptional regulation of pcaIJF, we determined the transcriptional start site of the operon by using primer extension analysis. Two different primers were used to map the start site, and each yielded a single extension product only with RNA derived from cells grown in the presence of protocatechuate (Fig. 4A). The transcriptional start site of the pcaIJF operon mapped to an A residue located 20 nucleotides upstream of the predicted PcaI start codon. Figure 4B shows the sequence of the pcaI promoter, including the inferred −10 and −35 hexanucleotide regions. Examination of the promoter region revealed the presence of a 9-bp palindromic sequence (5′ GCTTATTCG 3′) coincident with the predicted −10 region. Comparison of pcaIp with other S. meliloti promoters reveals some sequence similarity, particularly with respect to the inferred −35 regions (1, 2, 11, 26, 27, 32).
FIG. 4.
Analysis of the pcaI promoter in S. meliloti. (A) RNA isolated from S. meliloti wild-type derivative strain Rm1021 was used in primer extension reactions. Lane 1, no extension products were observed using RNA obtained from cells grown in the absence of protocatechuate; lane 2, a single extension product was observed using RNA derived from cells grown in the presence of protocatechuate. Extension reactions were performed using two different primers; however, only results obtained with one of the two primers are shown (reactions with both primers identified the same start site). Sequencing reactions were performed using the same primer used in extension reactions, and results are shown to the left of the extension reaction products. The arrows indicate the extension product (right) and identify the corresponding nucleotide (left). (B) pcaI promoter region, including the inferred −10 and −35 regions (underlined). The transcriptional start site is identified (in bold, enlarged font), and the start codon is also indicated (enlarged font at far right). (C) Comparison of the pcaI promoter with other previously determined promoters in S. meliloti.
Regulation of the pcaIJF operon requires an IclR-type regulator encoded by SMb20586.
The regulatory systems associated with the protocatechuate branch of the β-ketoadipate pathway have been described for a few species. In P. putida, expression of pca genes (with the exception of pcaHG) is regulated by the IclR-type transcriptional regulator PcaR, with β-ketoadipate serving as an inducing metabolite (17, 34). In A. baylyi, expression of the pcaIJFBDKCHG operon is regulated by the IclR-type protein PcaU (14), which acts as both a repressor and an activator of the operon, depending upon the presence of the inducer protocatechuate (51). In A. tumefaciens and Rhizobium leguminosarum, β-ketoadipate succinyl-CoA transferase activity is likewise induced by β-ketoadipate, and in A. tumefaciens, expression of pcaIJ is subject to regulation by an IclR-type protein (36, 39).
In S. meliloti, the pcaIJF operon is located beside a gene (SMb20586) encoding a putative IclR-type protein. To determine whether this putative regulator is involved in the regulation of pcaIJF gene expression, a streptomycin/spectinomycin antibiotic cassette was used to inactivate SMb20586, generating strain RmK1014 (as verified by Southern hybridization). RmK1014 was unable to grow with protocatechuate but grew like the wild type with glycerol or glucose as the sole carbon source, showing that SMb20586 is essential for protocatechuate metabolism (data not shown).
A promoterless gusA reporter gene was inserted at the 3′ end of the pcaIJF operon in the genome in order to monitor expression of this operon. The fusion was designed such that pcaF was not disrupted, as was verified by growth of the fusion strain with protocatechuate as a sole carbon source. In the wild-type Rm1021 background, expression of pcaF::gusA increased 10-fold and 5-fold following growth in the presence of protocatechuate and adipate, respectively (Table 4). In this instance, adipate was used as an analogue of β-ketoadipate. In contrast, expression of pcaF::gusA in the SMb20586 mutant background was minimal regardless of whether protocatechuate or adipate was in the growth media. This demonstrated that a product encoded by SMb20586 was required for pcaIJF expression.
TABLE 4.
Expression of pcaF-gusA fusion in S. meliloti wild-type and PcaR− backgrounds
| Strain | Relevant genotype | Growth condition | β-Glucuronidase activitya (Miller units [SD]) |
|---|---|---|---|
| RmK1015 | Rm1021 pcaF::gusA | Glycerol | 27 (2.7) |
| Glycerol plus adipate | 131 (11.7) | ||
| Glycerol plus PCAb | 259 (18.6) | ||
| RmK1016 | Rm1021 pcaF::gusA pcaR::Ω | Glycerol | 52 (3.0) |
| Glycerol plus adipate | 48 (4.1) | ||
| Glycerol plus PCA | 66 (6.8) |
Shown are averages of values obtained from three independent cultures grown overnight in LBmc and subcultured into M9 minimal medium with 0.5% glycerol ± 5 mM protocatechuate or 30 mM adipate at 30°C for 4 h.
PCA, protocatechuate.
The regulator encoded by SMb20586 from S. meliloti shares 59% amino acid identity with PcaR of A. tumefaciens (53) and 41% identity with PcaR from P. putida (46). In P. putida and A. tumefaciens, adipate is utilized as an inducer analogue of β-ketoadipate (36, 40), and based on the inducing activity of adipate as revealed in Table 4, we conclude that β-ketoadipate is the in vivo metabolite responsible for pcaIJF expression in S. meliloti. Based upon its amino acid sequence similarity and role in the regulation of pcaIJF expression, we have renamed SMb20586 pcaR.
Transcriptional regulator PcaR participates in autoregulation.
To examine whether PcaR could also autoregulate its own synthesis, the region upstream of the pcaR translational start site was amplified and cloned into the gusA reporter vector pFus1 (45) to create pTH1335. This plasmid was then conjugated into S. meliloti strains Rm1021 and RmK1014 (a pcaR::Ω derivative), and promoter activity was monitored via β-glucuronidase enzyme assays (Table 5). In both strains, reporter enzyme activities are comparable in cells grown with and without protocatechuate, indicating that pcaR expression is not influenced by the presence of this compound. Expression of pcaR::gusA in the low-copy-number plasmid pTH1335 is relatively low (comparable to expression of the pcaF::gusA fusion in uninduced cells), as expected of a regulatory gene. However, expression from the pcaR promoter was increased fivefold in the pcaR mutant compared to that in Rm1021 (Table 5). These results demonstrate that PcaR expression is negatively autoregulated.
TABLE 5.
Expression of pcaR-gusA fusion in S. meliloti wild-type and PcaR− backgrounds
| Strain | Relevant genotype | Growth condition | β-Glucuronidase activitya (Miller units [SD]) |
|---|---|---|---|
| RmP892 | Rm1021 (pTH1335) | Glycerol | 62 (2.2) |
| Glycerol plus PCAb | 42 (2.6) | ||
| RmP893 | Rm1021 pcaR::Ω (pTH1335) | Glycerol | 291 (8.2) |
| Glycerol plus PCA | 261 (3.1) |
Shown are averages of values obtained from three independent cultures grown overnight in LBmc and subcultured into M9 minimal medium with 0.5% glycerol ± 5 mM protocatechuate at 30°C for 4 h.
PCA, protocatechuate.
Analysis of the β-ketoadipate pathways in S. meliloti and A. tumefaciens.
Many aspects regarding the organization and regulation of genes encoding enzymes involved in the upper portion of the β-ketoadipate pathway (metabolism of protocatechuate to β-ketoadipate) are conserved between S. meliloti and A. tumefaciens. In addition to amino acid sequence similarities between homologues of the two species (which range from 60 to 77% identity), the organizations of genes into a single operon (pcaDCHGB) are identical. Likewise, regulation of the operon in both species is mediated by a LysR-type transcriptional regulator. Identification of the S. meliloti pcaD transcriptional start site and comparison with M. loti and A. tumefaciens sequences reveal that the pcaD promoter is likely conserved among these species. Similarly, previous work utilizing an A. tumefaciens pcaD::lacZ fusion indicates that the PcaQ binding sites of this species are recognized by the S. meliloti homologue (38). Although the evidence is limited, it is also quite likely that β-carboxy-cis,cis-muconate and γ-carboxymuconolactone serve as coinducing metabolites required for the PcaQ-regulated expression of the pcaDCHGB operon in S. meliloti, as has been shown for A. tumefaciens.
With respect to genes whose products participate in the lower portion of the pathway (conversion of β-ketoadipate to succinate and acetyl-CoA), certain similarities between S. meliloti and A. tumefaciens may also be observed. In both species, genes encoding subunits of a β-ketoadipate succinyl-CoA transferase (pcaI and pcaJ) are organized into an operon whose expression is regulated by an IclR-type transcriptional regulator (PcaR), with β-ketoadipate serving as a coeffector. On the other hand, amino acid sequence identity between PcaIJ of S. meliloti and A. tumefaciens is quite low and signature sequences present in the A. tumefaciens PcaIJ are absent or modified in the S. meliloti protein.
Comparison of the β-ketoadipate pathways in S. meliloti and A. tumefaciens can be extended to include PobA (4-hydroxybenzoate hydroxylase), an enzyme involved in the catalysis of 4-hydroxybenzoate to protocatechuate. In S. meliloti, a gene annotated as pobA is situated between the two pca operons. Despite the presence of this gene, S. meliloti strain Rm1021 is unable to utilize 4-hydroxybenzoate as a sole carbon source and we were unable to isolate an Rm1021 mutant that acquired this capability (data not shown). Likewise, an auxanographic study of Rhizobiaceae reported S. meliloti incapable of growing upon this compound (41). Moreover, using two independent transcriptional fusions to the S. meliloti pobA gene (utilizing gfp and gusA as reporters), we detected only basal pobA expression which did not increase upon addition of 4-hydroxybenzoate or protocatechuate to the growth medium (data not shown). In contrast, A. tumefaciens is able to grow at the expense of 4-hydroxybenzoate (41), and pobA expression in this species is induced by 4-hydroxybenzoate via PobR (39). In A. tumefaciens (and R. leguminosarum), an AraC family transcriptional regulator (PobR) regulates pobA expression (39), but there are no araC homologues located nearby in the S. meliloti genome. A putative LysR-type regulator (SMb20582) is located directly downstream of pobA, and although members of this family of regulators are typically transcribed divergently from a target gene, it is possible that this gene encodes a pobA regulator. In A. baylyi, an IclR-type regulator positively regulates expression of the pobA gene (7); however, the only close IclR gene in S. meliloti has been identified as pcaR (this work) because it regulates pcaIJF expression. Expression levels of pobA (as determined by reporter enzyme assays) in wild-type and pcaR mutant backgrounds are comparable, and we have concluded that PcaR is not involved in the regulation of pobA gene expression. Consequently, the identity of a regulator of pobA expression in S. meliloti, if one exists, remains obscure.
The supraoperonic organization of genes whose products participate in the catabolism of protocatechuate and related compounds has been well documented (4, 18, 39). In A. tumefaciens and S. meliloti, the two pca operons are clustered in close proximity, flanking the putative pobA gene. It has been proposed that this supraoperonic organization in A. tumefaciens arose as the result of the acquisition of these genes as a unit and that the protocatechuate pathway evolved prior to the divergence of Agrobacterium and Rhizobium species (39). Although much of the genetic organization and regulation in these systems has been conserved, the differences observed (particularly) with respect to PcaIJ of S. meliloti and A. tumefaciens are inconsistent with a shared history. It may be that although the protocatechuate catabolic pathway was established prior to Agrobacterium and Rhizobium speciation, the pcaIJ genes present today were acquired at some point afterwards independently in one or both genera. Possibly, this punctuated assembly of the two pca operons resulted in the loss of the pobA regulator, leaving S. meliloti unable to efficiently metabolize 4-hydroxybenzoate.
Acknowledgments
This work was supported by grants to T.M.F. from the Natural Sciences and Engineering Council of Canada, Genome Canada through the Ontario Genomics Institute, and the Ontario Research and Development Challenge Fund.
REFERENCES
- 1.Albright, L. M., C. W. Ronson, B. T. Nixon, and F. M. Ausubel. 1989. Identification of a gene linked to Rhizobium meliloti ntrA whose product is homologous to a family to ATP-binding proteins. J. Bacteriol. 171:1932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, Y. M., E. Holmgren, and I. P. Crawford. 1989. Rhizobium meliloti anthranilate synthase gene: cloning, sequence, and expression in Escherichia coli. J. Bacteriol. 171:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondelet-Rouault, M., J. Weiser, A. Lebrihi, P. Branny, and J. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the π interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 4.Buchan, A., E. L. Neidle, and M. A. Moran. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cánovas, J. L., and R. Y. Stanier. 1967. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella calcoacetica. 1. General aspects. Eur. J. Biochem. 1:289-300. [DOI] [PubMed] [Google Scholar]
- 6.Deghmane, A. E., D. Giorgini, L. Maigre, and M. K. Taha. 2004. Analysis in vitro and in vivo of the transcriptional regulator CrgA of Neisseria meningitidis upon contact with target cells. Mol. Microbiol. 53:917-927. [DOI] [PubMed] [Google Scholar]
- 7.DiMarco, A. A., B. Averhoff, and L. N. Ornston. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durham, D. R., L. A. Stirling, L. N. Ornston, and J. J. Perry. 1980. Intergeneric evolutionary homology revealed by the study of protocatechuate 3,4-dioxygenase from Azotobacter vinelandii. Biochemistry 19:149-155. [DOI] [PubMed] [Google Scholar]
- 9.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorhölter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Pühler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, R. F., H. L. Brierley, J. T. Mulligan, and S. R. Long. 1987. Transcription of Rhizobium meliloti nodulation genes. Identification of a nodD transcription initiation site in vitro and in vivo. J. Biol. Chem. 262:6849-6855. [PubMed] [Google Scholar]
- 12.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 13.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F. J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 14.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göbel, M., K. Kassel-Cati, E. Schmidt, and W. Reineke. 2002. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: cloning, characterization, and analysis of sequences encoding 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. J. Bacteriol. 184:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goethals, K., M. Van Montagu, and M. Holsters. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood, C. S., N. N. Nichols, M. K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 19.Hoet, P. P., and R. Y. Stanier. 1970. Existence and functions of two enzymes with β-ketoadipate:succinyl-CoA transferase activity in Pseudomonas fluorescens. Eur. J. Biochem. 13:71-76. [DOI] [PubMed] [Google Scholar]
- 20.Holding, A. J., and J. G. Collee. 1971. Routine biochemical tests, p. 1-32. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology. Academic Press, London, United Kingdom.
- 21.Ieva, R., C. Alaimo, I. Delany, G. Spohn, R. Rappuoli, and V. Scarlato. 2005. CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription. J. Bacteriol. 187:3421-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 24.Kaschabek, S. R., B. Kuhn, D. Müller, E. Schmidt, and W. Reineke. 2002. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: purification and characterization of 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. J. Bacteriol. 184:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowalchuk, G. A., G. B. Hartnett, A. Benson, J. E. Houghton, K. L. Ngai, and L. N. Ornston. 1994. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene 146:23-30. [DOI] [PubMed] [Google Scholar]
- 26.Leong, S. A., P. H. Williams, and G. S. Ditta. 1985. Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 13:5965-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLellan, S. R., L. A. Smallbone, C. D. Sibley, and T. M. Finan. 2005. The expression of a novel antisense gene mediates incompatibility within the large repABC family of alpha-proteobacterial plasmids. Mol. Microbiol. 55:611-623. [DOI] [PubMed] [Google Scholar]
- 28.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 31.Ornston, L. N., and R. Y. Stanier. 1966. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. J. Biol. Chem. 241:3776-3786. [PubMed] [Google Scholar]
- 32.Østerås, M., B. T. Driscoll, and T. M. Finan. 1995. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J. Bacteriol. 177:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parales, R. E., and C. S. Harwood. 1992. Characterization of the genes encoding β-ketoadipate:succinyl-coenzyme A transferase in Pseudomonas putida. J. Bacteriol. 174:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parales, R. E., and C. S. Harwood. 1993. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 175:5829-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parke, D. 1993. Positive regulation of phenolic catabolism in Agrobacterium tumefaciens by the pcaQ gene in response to β-carboxy-cis,cis-muconate. J. Bacteriol. 175:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 177:3808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parke, D. 1996. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J. Bacteriol. 178:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parke, D. 1996. Conservation of PcaQ, a transcriptional activator of pca genes for catabolism of phenolic compounds, in Agrobacterium tumefaciens and Rhizobium species. J. Bacteriol. 178:3671-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parke, D. 1997. Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol. Lett. 146:3-12. [Google Scholar]
- 40.Parke, D., and L. N. Ornston. 1976. Constitutive synthesis of enzymes of the protocatechuate pathway and of the β-ketoadipate uptake system in mutant strains of Pseudomonas putida. J. Bacteriol. 126:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parke, D., and L. N. Ornston. 1984. Nutritional diversity of Rhizobiaceae revealed by auxanography. J. Gen. Microbiol. 130:1743-1750. [Google Scholar]
- 42.Parke, D., and L. N. Ornston. 1986. Enzymes of the β-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J. Bacteriol. 165:288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 44.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 45.Reeve, W. G., R. P. Tiwari, P. S. Worsley, M. J. Dilworth, A. R. Glenn, and J. G. Howieson. 1999. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology 145:1307-1316. [DOI] [PubMed] [Google Scholar]
- 46.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 48.Spaink, H. P., J. H. Robert, C. A. Okker, E. P. Wijffelman, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 49.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trautwein, G., and U. Gerischer. 2001. Effects exerted by transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wierenga, R. K., P. Terpstra, and W. G. Hol. 1986. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]
- 53.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 54.Yeh, W. K., and L. N. Ornston. 1981. Evolutionarily homologous α2β2 oligomeric structures in β-ketoadipate succinyl-CoA transferases from Acinetobacter calcoaceticus and Pseudomonas putida. J. Biol. Chem. 256:1565-1569. [PubMed] [Google Scholar]