Abstract
The metaSHARK (metabolic search and reconstruction kit) web server offers users an intuitive, fully interactive way to explore the KEGG metabolic network via a WWW browser. Metabolic reconstruction information for specific organisms, produced by our automated SHARKhunt tool or from other programs or genome annotations, may be uploaded to the website and overlaid on the generic network. Additional data from gene expression experiments can also be incorporated, allowing the visualization of differential gene expression in the context of the predicted metabolic network. metaSHARK is available at http://bioinformatics.leeds.ac.uk/shark/.
INTRODUCTION
Our expanding knowledge of the metabolic capabilities of a wide range of organisms, as derived from genome sequencing and metabolic reconstruction efforts, presents a need for new methods for effective visualization of metabolic networks. Navigation of these networks remains difficult for many researchers, compounded by the various levels at which biochemical pathways can be fractionated. Many online resources are now available for the study of metabolic networks on a genome scale [e.g. KEGG (1), BioCyc (2), PUMA2 (3), aMAZE (4) and Reactome (5)]. However, visualization of the network data at these websites is limited to a number of pre-defined static pathway diagrams. This approach neglects the potential variability of the structure of metabolic pathways between organisms, and makes the discovery of novel pathways difficult.
The metaSHARK (metabolic search and reconstruction kit) web server addresses this problem by providing an intuitive and flexible interface to the metabolic network data, called SHARKview. This runs as a Java applet in the user's web browser, and does not require the installation of any additional software. SHARKview visualizations of metabolic pathways are completely customizable, allowing biologists to explore the network neighbourhood of enzymes of interest and to formulate hypothetical routes for the synthesis or catabolism of particular compounds.
By registering for a free account on metaSHARK, users are able to upload their own metabolic reconstruction data to a password-protected area on the website. The SHARKview interface can then be used to explore the metabolic network associated with the enzymes that have been asserted in a particular species, or to compare reconstructions for two different species. These customized visualizations may be saved and printed. The updated metaSHARK server now also makes it possible to visualize gene expression datasets in the context of an organism's predicted metabolic network.
EXPLORING METABOLIC NETWORKS
The metaSHARK website
The metabolic network data currently used in metaSHARK are derived from the KEGG (LIGAND) database (1). Each enzyme, reaction and compound in LIGAND has a corresponding page in metaSHARK presenting its associated data and hyperlinks to KEGG. Enzyme pages also include links to the PRIAM resource for enzyme-specific protein sequence profiles (6), which forms the basis of our SHARKhunt tool for the detection of enzymes within genomic DNA sequence (7), now available to download for Windows, Linux and Mac OS X (Power PC) platforms.
From the metaSHARK homepage, users may register to receive notification of server updates and to receive their own accounts for storing pathway visualizations, metabolic reconstruction and gene expression data. Illustrations of the capabilities of the metaSHARK platform are shown on our server for the human malaria parasite, Plasmodium falciparum, using the published genome sequence (8) and publicly available expression data, downloaded from the PlasmoDB resource (9).
The SHARKview interface
The SHARKview representation of the metabolic network is made up of nodes and directed arcs of different types. Although at first glance this representation may appear unusual to the biologist or biochemist user, it offers many advantages over the traditional curved arrow notation in terms of network navigation and layout, and maps easily onto both the KEGG (1) and SBML (10) network representations.
The class of an object in the database is represented by the shape of its node: squares represent reactions, circles/ellipses represent compounds, and rounded rectangles represent enzymes. The relationships between network objects are represented by the arcs connecting the nodes. Arcs shown with open (chevron) arrowheads connect compounds with reactions. The direction of an arc shows whether a compound is a substrate (input) or product (output) of a reaction. Since a large number of metabolic reactions are reversible, the default direction of reversible reactions is the left-to-right direction taken from the KEGG equation. In cases where more than one molecule of a particular type is consumed or produced in the reaction, this is shown by a small number to one side of the arc. Arcs of a second type, with a solid triangular arrowhead, connect enzymes with the reactions that they catalyse.
The different classes of nodes may be labeled according to their names in KEGG, or by EC number (11) or Gene Ontology ID (12) in the case of enzymes.
Some chemicals involved in metabolism, such as ATP, water and NAD+, appear in so many different reactions that if they were represented in the same way as the other metabolites, the network view would quickly degenerate into a tangled mess. To overcome this problem, SHARKview differentiates between sparsely-connected ‘path’ metabolites and these highly-connected ‘pool’ metabolites. Pool metabolites are sometimes referred to as ‘ubiquitous metabolites’, because they are usually chemicals that can be considered to be present in excess throughout the cell. A pool metabolite is represented in SHARKview by multiple copies of a blue circle or ellipse, one attached to each reaction (square) in which it participates. Path metabolites (orange circles or ellipses in the default view) only appear once in the SHARKview diagram. Some of the chemicals that are generally considered to be pool metabolites have certain reactions and pathways in which they play a more central role. A good example of this is ATP, which appears in many reactions as an energy-providing cofactor, as well as being a structural component in the pathways of nucleotide synthesis. metaSHARK stores information about which compounds play the part of pool metabolites in each reaction, so that the compound is represented in SHARKview as a pool or path metabolite depending on its context. If necessary, the user may also change which compounds are considered as part of the pool.
SHARKview makes it easy for the user to navigate the metabolic network, to construct his or her own views of the pathways it contains, and to produce high-resolution PNG snapshots of a metabolic network visualization. The applet downloads only a small part of the network at a time, greatly improving performance whilst maximizing flexibility. The TouchGraph package (http://www.touchgraph.com/) is used in SHARKview to display the network interactively. As nodes are added to or removed from the display, TouchGraph dynamically alters the layout to accommodate the changes.
Visualizing metabolic reconstruction data
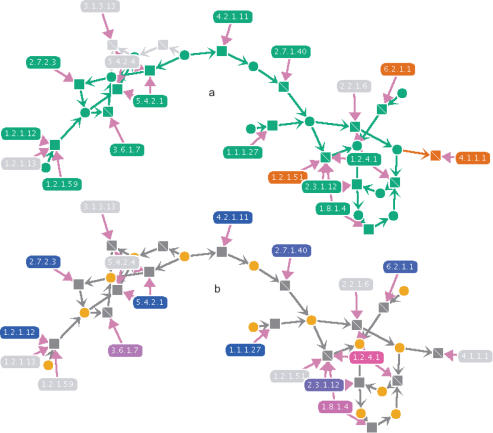
Metabolic reconstruction data produced by our automated tool, SHARKhunt (7), or derived from the output of other software such as PRIAM (6) or Pathway Tools (13), may be uploaded to the user's own password-protected area. Currently supported input formats include SHARKhunt XML output, plain lists of EC numbers (as output by PRIAM), and annotated EC lists including gene IDs and/or links to external web resources (see the metaSHARK website for further details). This information may be overlaid on the SHARKview network visualization to explore and compare the metabolic capabilities of different organisms (Figure 1a). In the case of SHARKhunt results, a colouring scheme based on an E-value score shows the degree of confidence for the presence of each specific enzymatic function within the genome sequence analysed. Links from SHARKview back to the main metaSHARK web pages enable users to inspect the sequence evidence for each enzyme (for SHARKhunt output), BLAST search the sequence, or explore external annotation resources.
Figure 1.
(a) A section of the glycolysis pathway from an automated metabolic reconstruction of the human malaria parasite P.falciparum, visualized using SHARKview. Metabolites are represented as circles, reactions as squares and enzymes as round-edged rectangles. Directed arcs between nodes show the effect of a reaction as the consumption and production of different metabolites. Nodes in green show reactions catalysed by enzymes for which good evidence has been found in the P.falciparum genome. Nodes in red show reactions catalysed by enzymes for which only tentative evidence has been found. Grey nodes show that no evidence for a catalysing enzyme was found. (b) The same pathway section for P.falciparum, with the enzyme nodes coloured according to their level of co-expression with lactate dehydrogenase (EC 1.1.1.27). Blue nodes show a high positive co-expression, whereas a colour towards pink shows a high negative co-expression.
Uploading gene expression data into metaSHARK
The metaSHARK website can now be used to view gene expression microarray data in the context of metabolic networks. To allow the data to be mapped onto the network, a gene expression dataset must be associated with a previously uploaded metabolic reconstruction. If this has been uploaded in the form of a list of EC numbers mapped to Gene IDs, then the gene expression data can be uploaded as a simple table, and the probe IDs will be automatically matched to the gene IDs in the reconstruction. If the reconstruction data has been produced with SHARKhunt, then the positions of the probes in the genome need to be included with the data so that they can be matched with the correct gene predictions. A script to combine the gene expression data and the probe positions, as well as example files can be found on the website. The acceptable format for the data is a table with the rows as individual genes or probes, and the columns as conditions. The expression values can either be single intensity values or expression fold changes. Owing to the different formats of data allowed, the expression of multiple probes cannot be combined into a single gene, and the data must be preprocessed. Once the data has been uploaded the expression levels for each enzyme individually can be displayed along with the reconstruction data on the website.
Visualizing gene expression data in SHARKview
The network visualization can show either the individual expression levels for each enzyme in the network, or the co-expression of the enzymes, calculated using the Pearson correlation coefficient. Owing to the variety of different data formats, cut-offs can be specified that define high, medium or low expression levels for individual enzymes, allowing the nodes to be coloured in a similar manner to the network reconstruction data. Alternatively, the enzymes can be coloured according to their co-expression with a selected enzyme in the network. For two genes a and b that have expression levels defined over N conditions, the co-expression is calculated using the Pearson correlation coefficient, defined as
where ai and bi are the gene expression signal values on array i, and bars are used to indicate the mean value for a gene's expression over all arrays. As the Pearson correlation score can be between −1 (for perfect negative correlation) and 1 (for perfect positive correlation), the nodes are coloured according to a gradient (Figure 1b). Alternatively, if there is a level of expression that is known to be significant then a user-defined cut-off can be entered. In the case where there is more than one gene in the dataset for a particular enzyme, one of these genes will automatically be chosen for display, and alternate genes can be selected manually.
CONCLUSIONS
The metaSHARK webserver provides an interactive visualization platform for the KEGG metabolic network in the form of the SHARKview applet. The generic network of compounds, reactions and enzymes may also be used as a framework for browsing metabolic reconstruction and gene expression datasets, shown by coloured nodes.
We expect metaSHARK to be useful to many researchers in generating hypotheses about metabolic function in particular species, and in suggesting avenues for further experimental investigations. The SHARKhunt tool for automated prediction of metabolic enzymes (downloadable from our website) may be applied to genomic DNA sequence to help kick-start metabolic reconstruction efforts, even before the appearance of a fully-annotated genome. Interactive browsing of the predicted network using SHARKview can greatly aid the discovery of novel pathways and new variants of known pathways. The direct comparison of metabolic reconstructions for two species may also provide useful information in drug target identification for pathogenic microbes.
Acknowledgments
The authors are grateful for funding for this project provided by the Medical Research Council (UK). J.W.P. is supported by a grant from the Biotechnology and Biological Sciences Research Council (UK). Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council (UK).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp P.D., Ouzounis C.A., Moore-Kochlacs C., Goldovsky L., Kaipa P., Ahren D., Tsoka S., Darzentas N., Kunin V., Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maltsev N., Glass E., Sulakhe D., Rodriguez A., Syed M.H., Bompada T., Zhang Y., D'Souza M. PUMA2—grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 2006;34:D369–D372. doi: 10.1093/nar/gkj095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemer C., Antezana E., Couche F., Fays F., Santolaria X., Janky R., Deville Y., Richelle J., Wodak S.J. The aMAZE LightBench: a web interface to a relational database of cellular processes. Nucleic Acids Res. 2004;32:D443–D448. doi: 10.1093/nar/gkh139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claudel-Renard C., Chevalet C., Faraut T., Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinney J.W., Shirley M.W., McConkey G.A., Westhead D.R. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 2005;33:1399–1409. doi: 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahl A., Brunk B., Crabtree J., Fraunholz M.J., Gajria B., Grant G.R., Ginsburg H., Gupta D., Kissinger J.C., Labo P., et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The Systems Biology Markup Language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 11.Enzyme Nomenclature. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes, NC-IUBMB. New York, NY: Academic Press; 1992. [Google Scholar]
- 12.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karp P., Paley S., Romero P. The Pathway Tools Software. Bioinformatics. 2002;18:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]