FIG. 1.
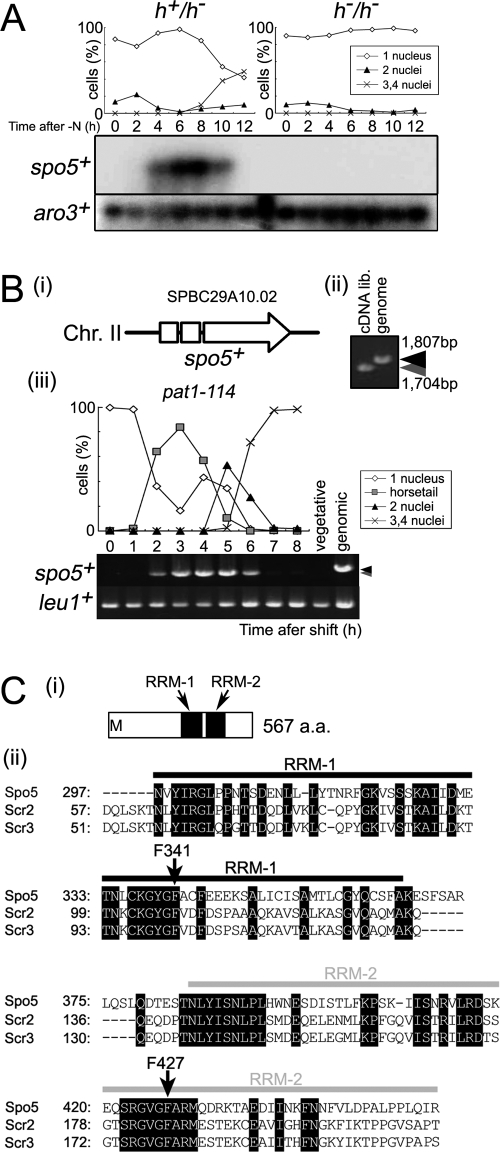
spo5+ is a meiosis-specific gene that encodes a protein with two RNA-binding motifs. (A) Northern blot analysis of spo5+ expression during meiosis. Total RNA was extracted from diploid h+/h− (CD16-1) and h−/h− (CD16-5) cells at the indicated times after they were subjected to nitrogen starvation, which induces CD16-1 but not CD16-5 cells to enter meiosis. Total RNA of these cells was blotted and probed with the protein-coding regions of the spo5+ and aro3+ (loading control) genes. The graph indicates the meiotic profiles of the cells used for RNA extraction. The frequencies of Hoechst 33342-stained cells with one, two, three, or four nuclei were assessed by counting at least 200 cells under a microscope. (B) PCR analysis of spo5+ transcripts during meiosis. (i) The structure of the spo5+ gene. The spo5+ gene consists of three exons which are shown by two open boxes and an open arrow. Chr., chromosome. (ii) The JZ670 (pat1-114) strain was induced to enter meiosis synchronously, and the total RNA from cells harvested at the indicated times was extracted and reverse transcribed to cDNA. PCR amplification of spo5+ was performed by using alternative cDNA from meiotic cells that were 3 h into meiosis, cDNA from vegetative cells, and genomic DNA as the template. PCR products were resolved on 1% agarose gels and stained by ethidium bromide. Black and gray arrowheads indicate the sizes of genomic spo5+ and the spliced spo5+ cDNA, respectively. lib., library. (iii) The difference in size of the spo5+ fragments amplified from genomic DNA or cDNA obtained 4 h into meiosis. (C) Schematic representation of the RRM in the RRM-bearing protein family. (i) The Spo5 protein has two RRMs in its C-terminal region as determined by SMART (http://smart.embl-heidelberg.de/). These motifs are designated as RRM-1 and RRM-2. a.a., amino acid. (ii) Alignment of RRM-1 and RRM-2 sequences with the RRM motif of Spo5, Scr2, and Scr3.