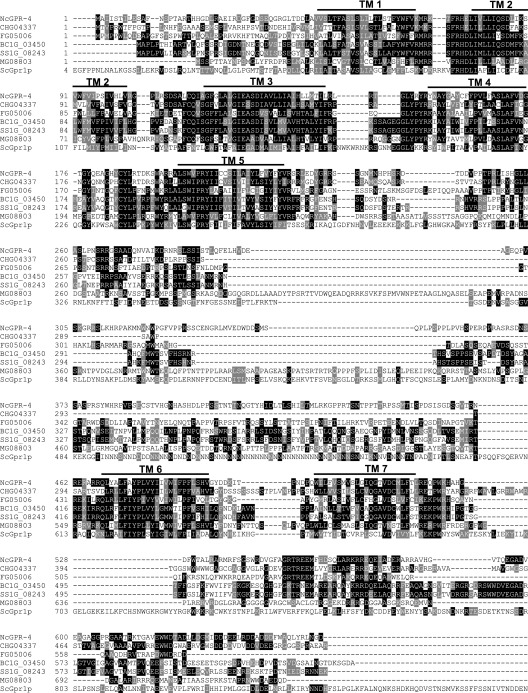
FIG. 1.
Alignment of GPR-4 (NcGPR-4) with homologous putative G-protein-coupled receptors from ascomycete fungi. The predicted amino acid sequences were aligned using ClustalW with shading by Boxshade (http://www.ch.embnet.org). The predicted transmembrane regions (TM1 to 7) are indicated by numbered lines above the sequences. Abbreviations: NcGPR-4, Neurospora crassa GPR-4 (accession no. NCU06312.1); ScGpr1p, Saccharomyces cerevisiae Gpr1p (accession no. JC5808); Mg08803, Magnaporthe grisea 70-15 hypothetical protein MG08803.4 (accession no. EAA51281); Fg05006, Gibberella zeae PH-1 (anamorph Fusarium graminearum) hypothetical protein FG05006.1 (accession no. XP_385182); CHG04337, Chaetomium globosum hypothetical protein CHG04337.1; SS1G_08243, Sclerotina sclerotiorum hypothetical protein SS1G_08243.1; BC1G_03450, Botrytis cinerea hypothetical protein BC1G_03450.1. An intron in FG05006.1 was misannotated, and the missing amino acid sequence LIMLLIYS was inserted between amino acids 79 and 80 in the original predicted protein sequence (http://www.broad.mit.edu/annotation/fgi/). Amino acids 121 to 137 were removed from the original predicted protein sequences of BC1G_03450.1 and SS1G_08243.1 (http://www.broad.mit.edu/annotation/fgi/), as these were misannotated and are actually part of an intron region (between amino acids 120 and 121 in this figure).