Abstract
About 5% of Clostridium perfringens type A isolates carry the cpe gene encoding the C. perfringens enterotoxin. Those cpe-positive type A isolates are important causes of food-poisoning and non-food-borne cases of diarrheas in humans, as well as certain veterinary cases of diarrhea. Previous studies have determined that the enterotoxigenic type A isolates causing both non-food-borne human gastrointestinal disease and veterinary disease carry their cpe genes on plasmids, while the type A isolates causing human food poisoning carry a chromosomal cpe gene. The present study reports on the successful development of a duplex PCR assay that can rapidly genotype enterotoxigenic type A isolates (i.e., determine whether those cpe-positive isolates carry a chromosomal or a plasmid-borne cpe gene). The availability of this rapid cpe genotyping assay capable of handling large numbers of samples provides a powerful new investigative tool for diagnostic, epidemiologic, and basic research purposes.
Isolates of the gram-positive spore former Clostridium perfringens are classified into five types (types A to E), depending upon their ability to express alpha, beta, epsilon, and iota toxins (16). Type A isolates, which predominate in the global C. perfringens population (11, 13), rank as important histotoxic and enteric pathogens of humans and domestic animals. Approximately 5% of all type A isolates produce (11, 13) another biomedically important toxin, named C. perfringens enterotoxin (CPE). Those CPE-positive type A isolates are traditionally recognized as the cause of C. perfringens type A food poisoning, the third most commonly reported food-borne illness in the United States (15). More recent studies (1-6, 12, 18) have also identified CPE-positive type A strains as an underappreciated cause of up to 5 to 20% of all cases of non-food-borne human gastrointestinal (GI) disease. CPE-associated non-food-borne GI disease cases are strongly associated with antibiotic use and involve enteric disease symptoms more severe and persistent than those typically seen with C. perfringens type A food poisoning (1-6, 12, 18). Instead of food-borne transmission, CPE-associated non-food-borne GI diseases appear to spread person to person or via ingestion of environmental contaminants (1-6, 12, 14, 18). Finally, CPE-associated type A isolates are also responsible for certain veterinary cases of diarrhea (14, 19).
CPE-positive C. perfringens type A isolates have been shown to carry their enterotoxin gene (cpe) on either the chromosome or a plasmid (8, 9, 20). The cpe gene is present on the chromosomes of most (or all) type A food-poisoning isolates (8, 20) but resides on large, single- or low-copy-number plasmids in most (or all) type A non-food-borne human GI disease or veterinary isolates (8, 9, 20). The recent discovery of these specific cpe genotype-disease associations has spurred considerable clinical and epidemiologic interest in the genotyping of disease-associated food or fecal cpe-positive C. perfringens type A isolates to determine whether they carry a chromosomal or a plasmid-borne cpe gene.
To date, cpe genotyping assays are limited to cpe Southern blot analyses of C. perfringens DNA subjected to pulsed-field gel electrophoresis (PFGE) or restriction fragment length polymorphism (RFLP) analysis (8, 9, 20). Those Southern blot analysis-based cpe genotyping procedures are slow and technically challenging, require expensive equipment and considerable technical expertise, and can handle only limited numbers of samples.
To address the need for an improved cpe genotyping assay, the present study reports on the development of a new PCR-based assay that can rapidly and simply determine whether a cpe-positive type A isolate carries a chromosomal or a plasmid-borne cpe gene.
MATERIALS AND METHODS
C. perfringens strains.
This study analyzed 84 cpe-positive C. perfringens type A isolates that had previously been genotyped for determination of the location of their cpe genes; i.e., their cpe genes had been localized to the chromosome or a plasmid by cpe Southern blot analysis-based RFLP or PFGE genotyping approaches (8, 17, 20). Those isolates tested included 57 isolates carrying plasmid-borne cpe genes from the following sources: 7 isolates from human antibiotic-associated diarrhea cases in the United Kingdom in the 1980s, 10 isolates from human sporadic diarrhea cases in the United Kingdom in the 1990s, 29 isolates from human antibiotic-associated diarrhea cases occurring in several different hospitals in the United States or Canada in the 1990s, 7 isolates from canine diarrhea cases in the United States in the 1990s, 1 isolate from a porcine enteritis case in the United Kingdom in the 1990s, 1 isolate from the peritoneal fluid of a canine in the 1990s, 1 fecal isolate from a nonsymptomatic dog in the United States in the 1990s, and 1 fecal isolate from a nonsymptomatic human in Japan in 2000.
Also examined were 27 type A isolates with chromosomal cpe obtained from the following sources: 8 isolates from human food-poisoning cases in the United Kingdom in the 1950s, 2 isolates from human food-poisoning outbreaks in the United States in the 1960s, 8 isolates from two different human food-poisoning outbreaks in the United States in the 1980s, 6 isolates from two different food-poisoning outbreaks in the United States in the 1990s, and 1 isolate from a porcine enteritis case in the United Kingdom in the 1990s.
As negative controls, this study also used ATCC 3624 and ATCC 297442, two C. perfringens type A isolates previously shown to be cpe negative (10, 17).
DNA isolation.
Starter vegetative cultures (10 ml) of each C. perfringens type A isolate were prepared by overnight growth at 37°C in fluid thioglycolate (FTG) broth (Difco). A 0.2-ml aliquot of each FTG broth culture was then inoculated into 10 ml of TGY broth (3% Trypticase, 2% glucose, 1% yeast extract, 0.1% cysteine [10]), which was then incubated at 37°C overnight. Genomic DNA was extracted from each overnight TGY culture, as described previously (10), and then quantified by measuring the UV absorption at 280 and 260 nm with a Bio-Rad Smartspec spectrophotometer. Aliquots of each extracted DNA preparation were then diluted to 10 ng/μl.
Single dcm-cpe PCR for detection of plasmid-borne cpe gene.
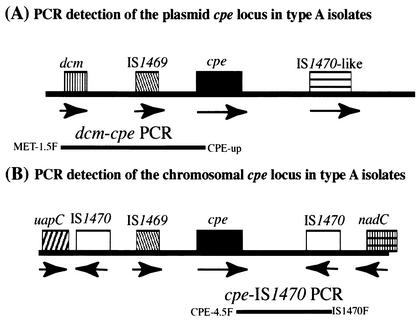
On the basis of recent results of studies describing the organization of the cpe locus in 13 type A isolates carrying a plasmid-borne cpe gene (17), the following primer pair was designed to amplify by PCR an ∼3.3-kb product from the apparently conserved DNA region (Fig. 1) between the upstream dcm sequences and the plasmid-borne cpe gene in type A isolates: 5′-CTCAGAGTTAGGAGCTAGCCCAACCC-3′ (primer MET-1.5F) and 5′-CCTAATATCCAACCATCTCC-3′ (primer CPE-up). The two primers were added (final concentration, 1 μM each) to a 50-μl PCR mixture containing 10 ng of purified DNA extracted from each C. perfringens type A isolate, 0.2 μM deoxynucleotide triphosphates (Promega), 5 μl of 10× Advantage 2 PCR buffer (Clontech), and 1 μl of Advantage 2 polymerase mix (Clontech). Those PCR mixtures were placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1st cycle, 94°C for 5 min; 2nd to 34th cycles, 94°C for 30 s, 63°C for 60 s, and 68°C for 180 s; and 35th cycle, 94°C for 90 s, 60°C for 90 s, and 68°C for 7 min. Aliquots (20 μl) of each PCR sample were then electrophoresed on a 1.5% agarose gel and visualized by staining with ethidium bromide.
FIG. 1.
Duplex PCR strategy for amplifying products from the chromosomal cpe locus and the plasmid-borne cpe locus of C. perfringens type A isolates. (A) Organization of the plasmid cpe locus of type A isolates (17); the bar underlying the open reading frames depicts the dcm-cpe PCR product specifically amplified from the plasmid cpe locus by use of the MET-1.5F and CPE-up primer pair. (B) Organization of the chromosomal cpe locus of type A isolates (17); the bar underlying the open reading frames depicts the cpe-IS1470 PCR product specifically amplified from the chromosomal cpe locus by use of the CPE-4.5F and IS1470F primer pair. Arrows indicate the orientations of the open reading frames.
Single cpe-IS1470 PCR for detection of chromosomal cpe gene.
On the basis of recent results of studies describing the organization of the cpe locus in five type A food-poisoning isolates carrying a chromosomal cpe gene (7, 17), the following primer pair was designed to PCR amplify an ∼2.1-kb product from the apparently conserved DNA region (see Fig. 1) between the chromosomal cpe gene and downstream IS1470 sequences in type A isolates: 5′-CAGTCCTTAGGTGATGGA-3′ (primer CPE-4.5F) and 5′-AACTAAATAGGCCTATAAATACC-3′ (primer IS1470F). The two primers were added (final concentration, 1 μM each) to a 50-μl PCR mixture containing the same concentrations of template and reagents described above for the dcm-cpe PCR. Those reaction mixtures were then subjected to similar PCR amplification conditions as described above for the dcm-cpe PCR. The resultant PCR products were visualized by ethidium bromide staining after electrophoresis on a 1.5% agarose gel.
Duplex PCR assay for genotyping cpe-positive C. perfringens type A isolates.
The dcm-cpe and cpe-IS1470 primer pairs (described above) were combined to create a duplex PCR genotyping assay capable of distinguishing between type A isolates carrying a chromosomal and a plasmid-borne cpe gene. The optimal final concentrations of the primers for this duplex PCR assay were determined by checkerboard titration (data not shown) to be 0.6 μM each for MET-1.5F and CPE-up and 0.8 μM each for CPE-4.5F and IS1470F. The duplex PCR was performed in a total volume of 50 μl under the same amplification conditions and with the same concentrations of purified DNA template and reagents described above for the two single PCRs for the amplification of sequences from either the type A chromosomal cpe locus or the type A plasmid-borne cpe locus. The PCR amplification conditions were also the same as those used for the single dcm-cpe and cpe-IS1470 PCRs. Duplex PCR products were detected by ethidium bromide staining after electrophoresis on a 1.5% agarose gel.
Duplex PCR cpe genotyping assay with lysates from C. perfringens colonies.
To test whether the duplex PCR cpe genotyping assay can use crude C. perfringens colony lysates as a template DNA source, a 0.1-ml aliquot of a cooked meat medium stock culture was transferred to 10 ml of FTG broth, which was then heat shocked at 72°C for 20 min before being incubated overnight at 37°C. A 0.2-ml aliquot of that FTG broth starter culture was inoculated into a fresh tube of FTG broth, which was incubated overnight at 37°C. A loopful of that overnight culture was then streaked onto a brain heart infusion (BHI) agar plate, which was then incubated for ∼18 h at 37°C in an anaerobic jar (BBL). About four to five colonies were taken from each BHI agar plate and suspended in 200 μl of sterile phosphate-buffered saline (PBS). Those suspended cells were microcentrifuged at 14,000 × g for 5 min, and the resultant pellet was washed twice with PBS prior to resuspension in 200 μl of PCR-grade H2O (Sigma). The microcentrifuge tube containing the resuspended cells was sealed with Parafilm and then microwaved (700 W) for a total of 20 min (administered as four 5-min heating treatments with 1-min cooling intervals) to induce bacterial lysis. The resultant lysate was cleared by microcentrifugation at 14,000 × g for 5 min, and 10 μl of that supernatant was used as template DNA for the duplex cpe genotyping PCR assay, as described above.
RESULTS
Development of a single dcm-cpe PCR assay for specific identification of C. perfringens type A isolates carrying a plasmid-borne cpe gene.
A recent study comparing the organization of the plasmid-borne and the chromosomal cpe loci in C. perfringens type A isolates (17) had determined that dcm sequences are present ∼3 kb upstream of the cpe gene in all 13 examined type A isolates carrying a plasmid-borne cpe gene (Fig. 1). However, dcm sequences were not detected in any of the five examined type A isolates carrying a chromosomal cpe gene (Fig. 1). Those previous observations suggested that primers specific for dcm and cpe sequences might be useful for the development of a PCR that could amplify a product (of ∼3.3 kb) from type A isolates carrying a plasmid-borne cpe gene but not from type A isolates carrying a chromosomal cpe gene.
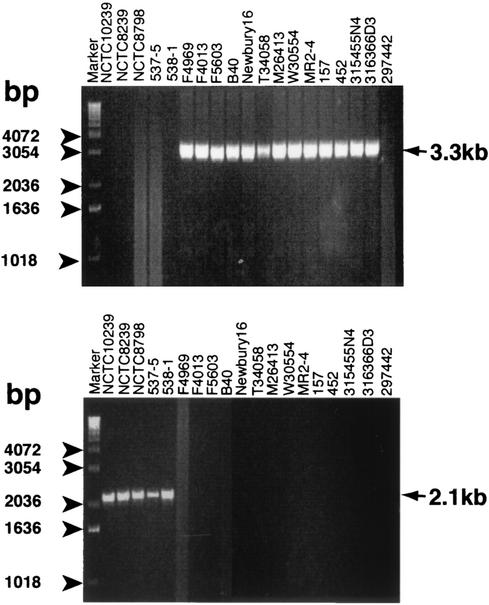
This hypothesis was tested by using purified DNA extracted from one cpe-negative isolate and the same 18 cpe-positive type A isolates examined in the previous study. As indicated by the representative results shown in Fig. 2A, the expected ∼3.3-kb PCR product was amplified by using purified DNA extracted from all 13 type A isolates whose plasmid-borne cpe loci had previously been investigated (17). However, no PCR product was amplified by using purified DNA extracted from any of the five previously examined type A isolates carrying a chromosomal cpe gene or from ATCC 297442, a type A isolate previously shown to be cpe negative (17).
FIG. 2.
Development of single PCR for identifying the plasmid-borne cpe locus or the chromosomal cpe locus of C. perfringens type A isolates. (A) Representative results obtained by the single dcm-cpe PCR (the expected product of 3.3 kb is indicated) with primers MET-1.5F and CPE-up. (B) Representative results obtained by the single cpe-IS1470 PCR (the expected product of 2.1 kb is indicated) with primers CPE-4.5F and IS1470F. The numbers on the left of each gel indicate the migration of molecular size markers.
Development of a single cpe -IS1470 PCR assay for specific identification of C. perfringens type A isolates carrying a chromosomal cpe gene.
The recent study comparing the organization of the plasmid-borne and chromosomal cpe loci in C. perfringens type A isolates (17) also determined that IS1470 sequences are present ∼2 kb downstream of the cpe gene in the five type A isolates carrying a chromosomal cpe gene examined previously. However, similarly oriented IS1470 sequences were not detected in the plasmid-borne cpe locus of the 13 type A isolates examined (Fig. 1). Those observations suggested that primers specific for IS1470 and cpe sequences might be useful for the development of a PCR that could amplify a product (of ∼2.1 kb) from type A isolates carrying a chromosomal cpe gene but not from type A isolates carrying a plasmid-borne cpe gene.
This hypothesis was tested by using purified DNA extracted from the same 18 well-characterized cpe-positive type A isolates and a single cpe-negative type A control isolate in the dcm-cpe PCR assay (Fig. 2A). As indicated by the representative results shown in Fig. 2B, the single cpe-IS1470 PCR amplified the expected ∼2.1-kb PCR product when purified DNA extracted from all five type A isolates whose chromosomal cpe locus had previously been investigated were used (17). However, this PCR failed to amplify a product when purified DNA extracted from any of the 13 previously examined type A isolates carrying a plasmid-borne cpe gene or the cpe-negative type A isolate, ATCC 297442, was used.
Development of a duplex PCR assay for genotyping cpe-positive C. perfringens type A isolates.
The success of the individual dcm-cpe and cpe-IS1470 PCR assays in identifying C. perfringens type A isolates carrying a plasmid-borne or a chromosomal cpe gene, respectively, suggested that those two PCRs could be combined in a duplex PCR assay capable of genotyping cpe-positive type A isolates, i.e., determining whether those isolates carry a plasmid-borne or a chromosomal cpe gene.
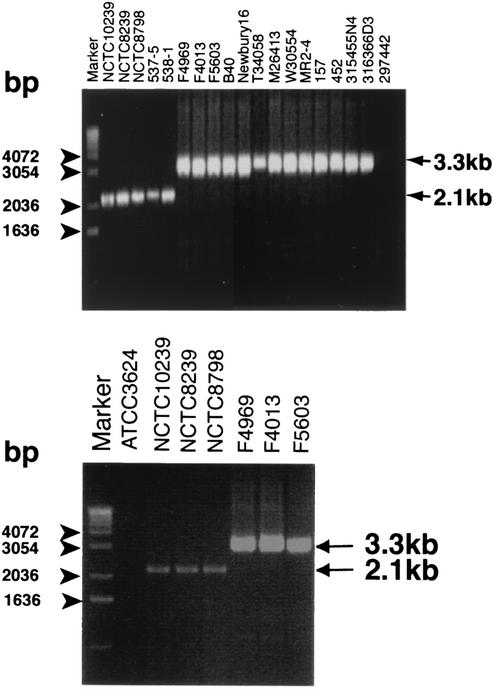
This hypothesis was first tested by using as a PCR template purified DNA extracted from the same 18 cpe-positive and 1 cpe-negative C. perfringens type A isolates (Fig. 2A and B). As indicated by the results shown in Fig. 3A, the duplex PCR assay correctly amplified an ∼3.3-kb product from all 13 initially tested type A isolates carrying a plasmid-borne cpe gene, an ∼2.1-kb product from all 5 initially tested type A isolates carrying a chromosomal cpe gene, and no product from the 1 initially tested cpe-negative type A isolate.
FIG. 3.
Development of a duplex PCR cpe genotyping assay for distinguishing C. perfringens type A isolates carrying a plasmid-borne cpe gene from isolates carrying a chromosomal cpe gene. (A) Representative duplex PCR results obtained by use of purified DNA extracted from isolates with chromosomal cpe genes (first 5 lanes from the left) or isolates carrying plasmid-borne cpe genes (the next 13 lanes). Also shown are the results of duplex PCR with purified DNA from cpe-negative isolate ATCC 297442. (B) Representative duplex PCR results obtained with colony lysates as a DNA template source. Results shown are for cpe-negative isolate ATCC 3624 (second lane from the left), three type A isolates with chromosomal cpe genes (next three lanes), and three type A isolates carrying plasmid-borne cpe genes (three lanes at the right). Arrows on the right in both panels depict the migration of the expected dcm-cpe PCR products (3.3 kb) and the expected cpe-IS1470 PCR products (2.1 kb). The numbers on the left of each gel indicate the migration of molecular size markers.
Given these promising results, further testing of the duplex PCR cpe genotyping assay was performed to confirm its reliability. For this purpose, purified DNA was extracted from 1 additional cpe-negative type A isolate and 66 additional cpe-positive type A isolates. Those 66 additional cpe-positive type A isolates had all been previously genotyped by traditional Southern blot analysis-based assays to determine whether they carry a chromosomal or plasmid-borne cpe gene (see Materials and Methods). However, the organization of the cpe locus in these 66 isolates had not yet been examined; i.e., it was unknown whether the isolates in this additional isolate group carrying plasmid-borne cpe genes had dcm sequences upstream of their cpe genes or whether the isolates in this additional isolate group with a chromosomal cpe gene had IS1470 sequences, in the same orientation as shown in Fig. 1, downstream of their cpe genes. Among these 66 additional cpe-positive type A isolates were 19 more food-poisoning isolates carrying a chromosomal cpe gene and 47 more human non-food-borne GI disease isolates or veterinary isolates carrying a plasmid-borne cpe gene.
In this testing of additional isolates, the duplex PCR assay amplified (data not shown) an ∼3.3-kb product from all 47 of the additional type A isolates carrying a plasmid-borne cpe gene, an ∼2.1-kb product from all 19 of the additional food-poisoning isolates carrying a chromosomal cpe gene tested, and no product from ATCC 3624, the additional cpe-negative type A isolate.
Finally, when the duplex PCR was tested by using a mock sample spiked with both the plasmid-borne and chromosomal copies of cpe, no PCR product was obtained. However, this failure is not a significant limitation of the assay, as none of the >84 natural CPE-positive type A isolates previously genotyped by RFLP or PFGE techniques had been found to simultaneously carry both chromosomal and plasmid cpe genes (8, 9, 20).
Use of the duplex PCR cpe genotyping assay with colony lysates.
The final experiment performed in this study involved testing of whether the duplex PCR cpe genotyping assay can use colony lysates as a DNA template source. For this analysis, 33 isolates carrying plasmid-borne cpe genes and 25 isolates with chromosomal cpe genes were randomly selected from our collection of 84 cpe-positive isolates whose purified DNA had been successfully genotyped by the duplex PCR in the studies whose results are shown in Fig. 3A. Representative results with colony lysates as a DNA template source are shown in Fig. 3B, which demonstrates that colony lysates from all three isolates with chromosomal cpe genes and all three isolates carrying plasmid-borne cpe genes, but not from the one cpe-negative isolate (ATCC 3624), supported the successful amplification of the appropriate PCR product in the duplex PCR cpe genotyping assay. Similarly, colony lysates from 49 other cpe-positive isolates tested also proved to be adequate template sources for this duplex PCR cpe genotyping assay. However, despite repeated attempts, no PCR product could be obtained with colony lysates from two other isolates carrying plasmid-borne cpe genes or one other isolate with a chromosomal cpe gene, even though those isolates had been successfully genotyped by the duplex PCR assay using their purified DNA as the template.
DISCUSSION
Prior to the current study, cpe-positive type A isolates of C. perfringens could be cpe genotyped only by Southern blotting of their DNA after PFGE or RFLP analysis. The successful development of the duplex PCR assay reported in this study represents a substantial improvement upon those existing cpe genotyping assays. For example, while Southern blots of RFLP or PFGE gels require 1 to 2 weeks for genotyping of cpe-positive type A isolates, our new PCR-based assay can determine whether a cpe-positive type A isolate carries a chromosomal or a plasmid-borne cpe gene in 1 to 2 days by use of purified DNA. By use of colony lysates as the template DNA source, the duplex PCR assay generates genotyping results within 6 h. Furthermore, our survey results with 55 cpe-positive type A isolates indicate that colony lysates suffice as a duplex PCR DNA template source for ∼95% of all cpe-positive type A isolates. In addition, the new duplex PCR assay more easily accommodates genotyping of large numbers of samples than Southern blotting of cpe on RFLP and PFGE gels does. Finally, this new genotyping assay requires no more sophisticated or costly equipment than a PCR thermocycler and agarose electrophoresis capability and uses PCR expertise already familiar to most laboratories.
The availability of this new PCR-based cpe genotyping assay provides a powerful tool for the study of the epidemiology of CPE-associated food-borne and non-food-borne human or veterinary GI diseases. Although CPE-positive type A isolates have been linked to enteric illness for many years, little is known about the natural reservoirs for the type A isolates carrying either plasmid-borne or chromosomal cpe genes. Similarly, it is unclear how or when type A isolates with chromosomal cpe genes enter the food chain to contaminate foods.
In addition, the new duplex PCR genotyping assay developed in this study holds considerable diagnostic promise. It should be useful to clinical microbiologists for differentiating human cases of CPE-associated food poisoning (which involve type A isolates with chromosomal cpe genes) and cases of CPE-associated non-food-borne human GI disease (which involve type A isolates carrying plasmid-borne cpe genes). It should also make routine testing for the involvement of isolates carrying plasmid-borne cpe genes in antibiotic-associated diarrhea cases much more feasible.
Finally, our results validating this duplex PCR assay are also informative regarding the diversity of cpe-positive type A isolates. Recent studies that have used Southern blotting, PCR, and sequencing approaches (17) demonstrated differences between the organization of the plasmid-borne cpe locus and the chromosomal cpe locus in 18 cpe-positive type A isolates. In the present study, our new duplex PCR assay assessed the cpe locus organization in approximately fourfold more cpe-positive type A isolates than had previously been examined. It is notable that these 66 additional cpe-positive type A isolates studied came from various geographic, host, and disease sources and had isolation dates ranging from the 1950s to the late 1990s (see Materials and Methods). Duplex PCR results with these 66 additional isolates provide important data confirming that the organizational differences noted between the plasmid-borne and chromosomal cpe loci in a previous study (17) are highly conserved in most (if not all) cpe-positive type A isolates. Specifically, the new duplex PCR assay's amplification of an ∼3.3-kb PCR product from all 47 additionally tested type A isolates carrying a plasmid-borne cpe gene supports previous suggestions (17) that the region upstream of the cpe gene is conserved in most (or all) type A isolates carrying plasmid-borne cpe genes. However, it should be noted that a previous study (17) did identify some diversity in the region downstream of the cpe locus among type A isolates carrying plasmid-borne cpe genes, where either IS1470-like or IS1151-like sequences can be situated; that diversity downstream of the plasmid-borne cpe gene in type A isolates explains why we designed primers to amplify the upstream dcm-cpe region for identifying type A isolates carrying plasmid-borne cpe genes by a duplex PCR assay.
Similarly, the ability of the duplex PCR to amplify the expected ∼2.1-kb PCR product from all 19 additionally tested type A isolates carrying a chromosomal cpe gene supports previous suggestions (17) that the region downstream of the cpe gene is conserved among most (if not all) type A isolates carrying chromosomal cpe genes. Whether sequence variations occur downstream of the chromosomal cpe locus of type A isolates requires further study.
Acknowledgments
This research was generously supported by Public Health Service grant AI19844 from the National Institute of Allergy and Infectious Diseases and by grant 2001-02517 from the Ensuring Food Safety Research Program of the U.S. Department of Agriculture.
REFERENCES
- 1.Abrahao, C., R. Carman, H. Hahn, and O. Liesenfeld. 2001. Similar frequency of detection of Clostridium perfringens enterotoxin and Clostridium difficile toxins in patients with antibiotic-associated diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 20:676-677. [DOI] [PubMed] [Google Scholar]
- 2.Asha, N., and M. Wilcox. 2002. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhea. J. Med. Microbiol. 51:891-894. [DOI] [PubMed] [Google Scholar]
- 3.Borriello, S. P. 1995. Clostridial diseases of the gut. Clin. Infect. Dis. 20(Suppl. 2):S242-S250. [DOI] [PubMed] [Google Scholar]
- 4.Borriello, S. P., F. E. Barclay, A. R. Welch, M. F. Stringer, G. N. Watson, R. K. T. Williams, D. V. Seal, and K. Sullens. 1985. Epidemiology of diarrhea caused by enterotoxigenic Clostridium perfringens. J. Med. Microbiol. 20:363-372. [DOI] [PubMed] [Google Scholar]
- 5.Borriello, S. P., A. R. Welch, H. E. Larson, F. Barclay, M. F. Stringer, and B. A. Bartholomew. 1984. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic-associated diarrhea. Lancet i:305-307. [DOI] [PubMed] [Google Scholar]
- 6.Brett, M. M., J. C. Rodhouse, T. J. Donovan, G. M. Tebbut, and D. N. Hutchinson. 1992. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhea. J. Clin. Pathol. 45:609-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 8.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 10.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daube, G., P. Simon, B. Limbourg, C. Manteca, J. Mainil, and A. Kaeckenbeeck. 1996. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ɛ, ι, θ, μ and enterotoxin) and for sialidase. Am. J. Vet. Res. 57:496-501. [PubMed] [Google Scholar]
- 12.Jackson, S., D. Yip-Chuck, J. Clark, and M. Brodsky. 1986. Diagnostic importance of Clostridium perfringens enterotoxin analysis in recurring enteritis among the elderly, chronic care psychiatric patients. J. Clin. Microbiol. 23:748-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks, S., E. Kather, P. Kass, and A. Melli. 2002. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J. Vet. Intern. Med. 16:533-540. [DOI] [PubMed] [Google Scholar]
- 15.McClane, B. A. 2001. Clostridium perfringens, p. 351-372. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 16.McClane, B. A., and J. I. Rood. 2001. Clostridial toxins involved in human enteric and histotoxic infections, p. 169-209. In H. Bahl and P. Duerre (ed.), Clostridia: bio/technology and medical applications. Wiley-VCH, Weinheim, Germany.
- 17.Miyamoto, K., G. Chakrabarti, Y. Morino, and B. A. McClane. 2002. Organization of the plasmid cpe locus of Clostridium perfringens type A isolates. Infect. Immun. 70:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mpamugo, O., T. Donovan, and M. M. Brett. 1995. Enterotoxigenic Clostridium perfringens as a cause of sporadic cases of diarrhoea. J. Med. Microbiol. 43:442-445. [DOI] [PubMed] [Google Scholar]
- 19.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens isolates associated with gastrointestinal disease in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]