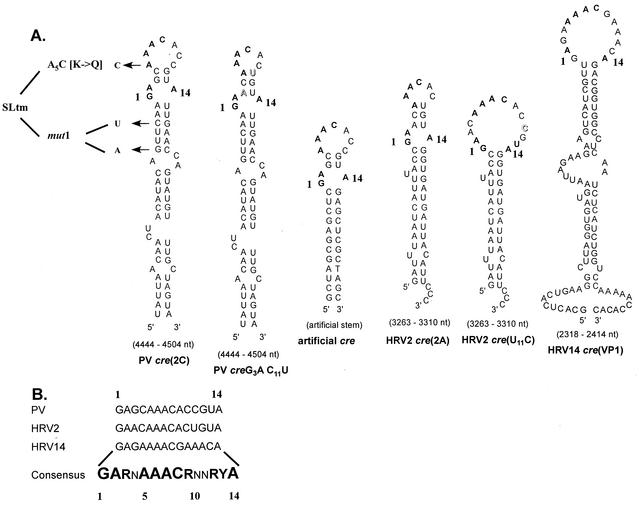
FIG. 2.
Comparison of predicted cre structures of various picornaviruses used in this study. (A) Hairpin structures predicted by the MFOLD RNA program (Michael Zuker) for wt PV-, HRV14-, HRV2-cre, HRV2-cre carrying a U11C mutation which enlarges the top loop structure, and a cre constructed to contain 14 nt from the PV-cre top loop and an unrelated artificial stem. Arrows, locations of the substitutions in mutant PV-cres corresponding to A5C, G4462A C4465U, and SLtm. Letters in shadow style, mutations introduced in PV-cre and HRV2-cre to alter their loop sizes. (B) Sequences of the top 14 nt of three naturally occurring picornavirus cres. The consensus sequence referred to in the text as the core sequence is in boldface. The degree of conservation at each nucleotide position is indicated by the size of the letter. N, any ribonucleotide; R, purine; Y, pyrimidine.