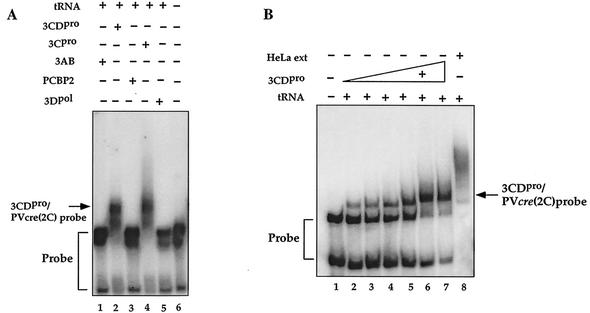
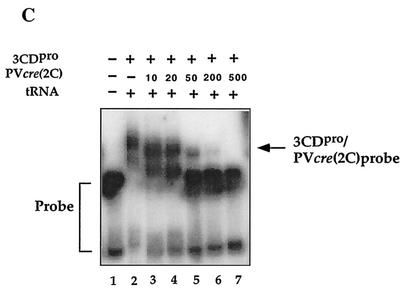
FIG. 8.
Relative abilities of poliovirus and HeLa cell proteins to form complexes with PV-cre(2C) RNA. The electrophoretic mobility shift assays were carried out with a 32P-labeled PV-cre(2C) probe in the presence of a 1,000-fold excess of tRNA as described in Materials and Methods. (A) Lane 6, radiolabeled PV-cre (63-nt, 7 nM) probe in binding buffer; lane 1, 3AB (0.8 μM); lane 2, 3CDpro (His tagged, 0.5 μM); lane 3, PCBP (1.1 μM); lane 4, 3Cpro (1.2 μM); lane 5, 3Dpol (1 μM). (B) Binding curve for PV-cre/3CDpro with increasing amounts of 3CDpro. Lane 1, PV-cre probe in binding buffer; lanes 2 to 7, probe in binding buffer with increasing amounts (0.05, 0.13, 0.2, 0.4, 0.5, and 0.6 μM) of 3CDpro (His tagged); lane 8, PV-cre probe in binding buffer and HeLa cell extract proteins (15 μg). (C) Competition for the binding of 3CDpro to the radiolabeled PV-cre probe (63 nt, 7 nM) with unlabeled PV-cre. Binding reactions were performed in the absence (lane 1) or in the presence (lanes 2 to 7) of recombinant 3CDpro (His tagged). Lanes 3 to 7 contain radiolabeled PV-cre probes in binding buffer, purified 3CDpro (0.5 μM; His tagged), and increasing concentrations (from 10 to 500 M excess) of unlabeled PV-cre RNA as the competitor.