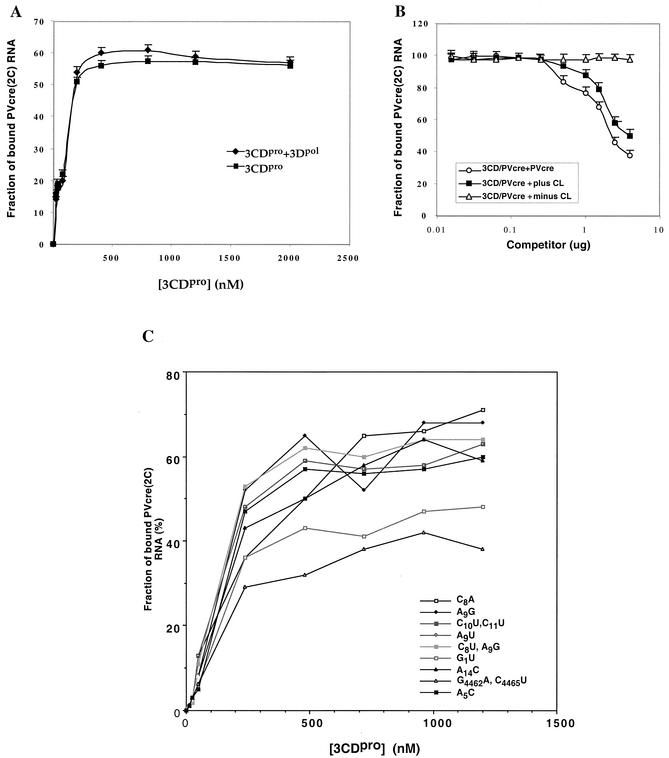
FIG. 9.
Filter binding assays to assess the effect of addition of 3Dpol or competitor (mutant PV-cre or cloverleaf) RNAs on the binding of 3CDpro to PV-cre(2C). (A) Binding reaction between PV-cre(2C) (63 nt, 7 nM) and 3CDpro (0.01 to 0.8 μM, untagged) performed in the absence (squares) or in the presence (diamonds) of 1 μM 3Dpol. 3CDpro used in this experiment and that described in the panel B legend is a recombinant (3Cpro/H40A) protein that does not carry additional tag sequences (23). The filter binding data collected are shown as the means of three measurements at each data point. (B) Effect of plus and minus strand poliovirus cloverleaf and wt PV-cre on the binding of 3CDpro to PV-cre. Competition was performed in the presence of either PV-cre (circles) or plus strand cloverleaf (squares) or minus strand cloverleaf (triangles) RNA competitors. Quantitative analysis of the 3CDpro/PV-cre complexes was carried out by using a filter-binding assay as described in Materials and Methods. (C) Comparison of the relative binding affinities of wt and mutant PV-cre RNAs for the site within 3CDpro in a filter-binding assay as described in Materials and Methods.