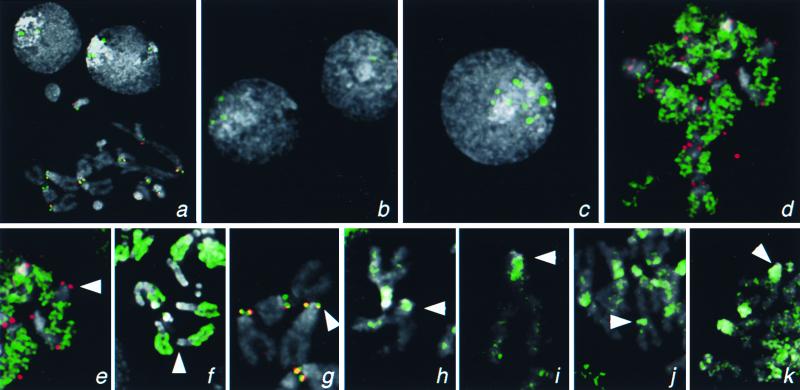
Figure 4.
Localization of H3-like protein-GFP fusions to centromeres of Drosophila. Kc167 cells were transfected with GFP fusion constructs driven by the cid promoter (a and d–k) or by basal (b) or induced (c) expression from the Hsp70 promoter. (a) Localization of Cid-GFP to the centromeres in a metaphase spread and to intense foci in interphase nuclei. Similar foci are seen in many interphase nuclei when Cid-GFP is expressed from the Hsp70 promoter (b and c) and are 10-fold more intense (185 ± 61 vs. 2,085 ± 979 pixel intensity) when this promoter has been induced (c). (d) Euchromatic labeling by cid-driven H3-GFP. Localization of GFP fusions on representative X chromosomes from mitotic figures with Drosophila H3 (e), Drosophila H2B (f), Drosophila Cid (g), human CENP-A (h), S. cerevisiae Cse4p (i), C. elegans HCP-3 (SWISS-PROT YMH3_CAEEL) (j), and C. elegans D6H3 (k) (GenBank accession no. AAB37052). GFP fluorescence was consistently lower with the H3-like heterolog-GFP fusion proteins than with the Cid- or the histone-GFP fusions. The X centromere is indicated with an arrowhead. The X chromosome is acrocentric, and the heterochromatin coheres well in metaphase spreads; thus, the position of the centromere can be identified by morphology alone. Identification of centromeres in metaphase spreads was confirmed by detection of the POLO kinetochore epitope. GFP signal is shown in green, the POLO epitope is shown in red (a, d, e, and g), and 4′,6-diamidino-2-phenylindole staining is shown in gray. Coincidence of GFP and POLO is yellow.