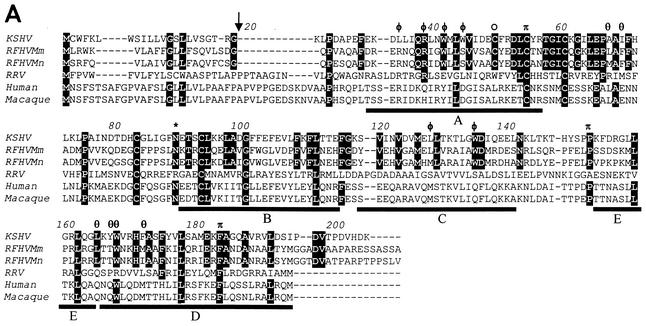
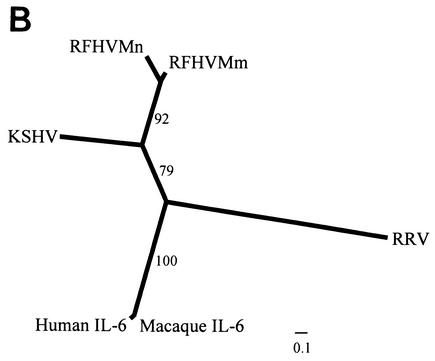
FIG. 6.
Comparison of viral and cellular IL-6 homologs. (A) Amino acid sequence alignment of the RFHVMm and RFHVMn vIL-6 homologs compared to the vIL-6 of KSHV and RRV and the human (NP_000591) and rhesus macaque (L26028) cellular IL-6. Amino acids conserved between the KSHV and RFHV sequences are highlighted; a potential N-linked glycosylation site and a cysteine residue conserved between KSHV and RFHV are indicated with an asterisk and an open circle, respectively. The predicted signal peptide cleavage site for the vIL-6 homologs is shown (arrow) and corresponds to the cleavage site determined for human IL-6. The four major alpha-helical regions (A, B, C, and D) and the minor helical region (E) determined for human IL-6 are indicated (47). The residues in sites II and III of KSHV vIL-6 which interact with the gp130 receptor subunit (see text) are indicated with φ and θ, respectively (7). The residues in site I which interact with the IL-6R subunit are indicated with π (23). The amino acid numbering is relative to the KSHV vIL-6 sequence. (B) Phylogenetic analysis of the alignment in A with the protein maximum-likelihood method. Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided.