FIG. 8.
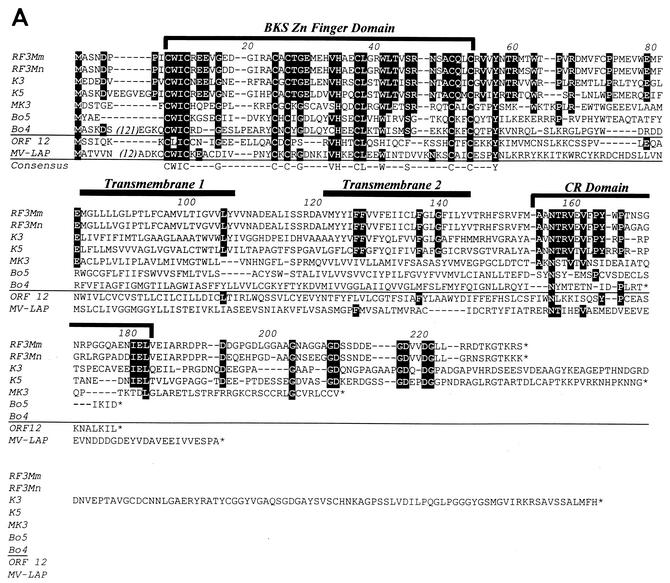
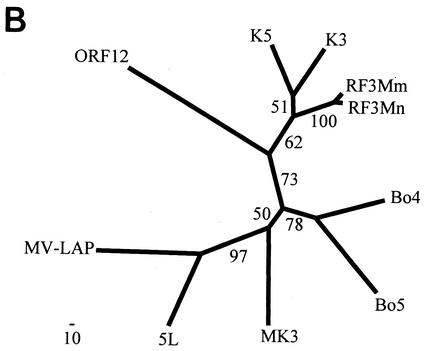
Comparison of ORF RF3 homologs. (A) Amino acid sequence alignment of the MIR homologs of KSHV (K3 and K5), RFHVMn (RF3Mn), RFHVMm (RF3Mm), BHV4 (Bo5 and Bo4), MHV68 (MK3), HVS (ORF 12), and myxoma virus (MV-LAP; AAK00734). The BKS zinc finger domain is indicated, with the positions of the hydrophobic transmembrane domains and the conserved region (CR) in the C-terminal domain shown. Residues identical within the K3, K5, RF3Mn, and RF3Mm sequences are highlighted. A consensus sequence for the BKS zinc finger domain is shown. (B) Phylogenetic analysis of the complete ORFs with the protein maximum-likelihood method with the addition of the 5L protein of yaba-like disease virus (NP_073390). Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided.