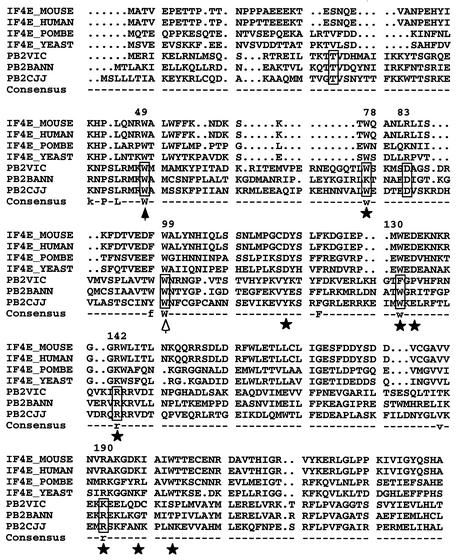
FIG. 1.
Alignment of PB2 and eIF4E protein sequences. The figure shows the alignment of PB2 proteins from influenza A, B, and C type viruses with a series of eIF4E proteins from various origins (POMBE, Schizosaccharomyces pombe; YEAST, Saccharomyces cerevisiae). The positions of PB2 protein included in boxes were mutagenized as indicated in the text. Stars mark conserved residues of eIF4E involved in interaction with the cap structure (40). The solid arrowhead indicates a position conserved in eIF4E whose mutation altered cap recognition, although it does not contact the cap structure. The open arrowhead indicates a position conserved in eIF4E but not involved in cap recognition. The numbers at the top of the sequence indicate the positions in the A/Victoria/3/75 sequence. The consensus line indicates residues conserved among PB2 and eIF4E sequences.