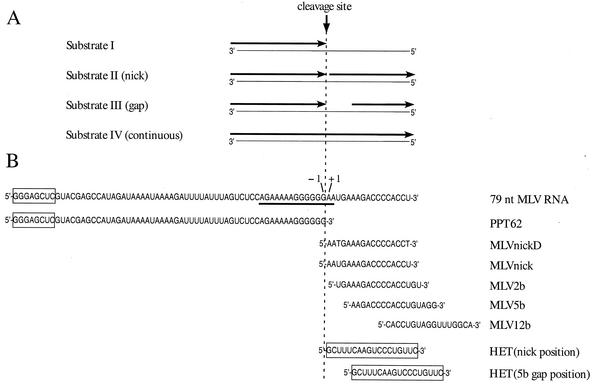
FIG. 1.
Model hybrid substrates with RNAs and oligonucleotides used in extension and cleavage analyses. (A) General structures of model hybrid substrates I to IV. Template strands are shown as thin lines with their 5′ and 3′ ends indicated. Oligonucleotides or RNAs annealed to template strands are shown as arrows originating at the 3′ ends. When applicable, positions of RNAs or oligonucleotides are described as upstream or downstream relative to the PPT primer cleavage site (indicated by vertical dashed line), which is based upon the plus-strand origin found within the PPT of the M-MuLV genome. Substrate I contains a 62-nt plus-strand primer (PPT62) with a 3′ end at position −1. Substrate II contains a downstream DNA or RNA oligonucleotide abutting the 3′ end of PPT62 and creating a nick. In substrate III, the 5′-end position of the downstream oligonucleotide creates a gap of 2, 5, or 12 bases from the 3′ end of PPT62. Substrate IV contains a 79-nt MLV RNA, which is a continuous RNA without a gap or nick. (B) The names and sequences of RNAs and oligonucleotides annealed to the template strands are shown relative to the −1 and +1 positions that border the plus-strand origin. The downstream oligonucleotides derived from the M-MuLV sequence are termed MLV, while the downstream oligonucleotide derived from an unrelated sequence is referred to as HET (for heterologous). The PPT sequence for the 79-nt MLV RNA is underlined. Sequences not derived from the M-MuLV genome are boxed. With the exception of the deoxyribonucleotide MLVnickD, all sequences are ribonucleotides.