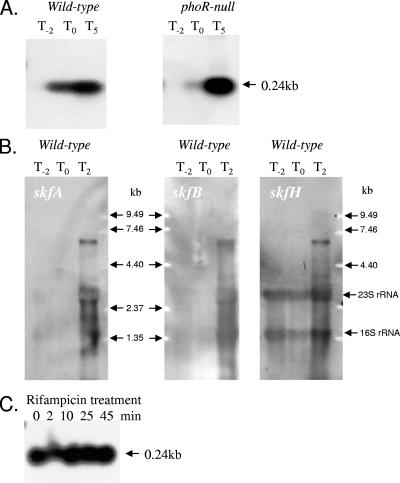
FIG. 2.
(A) Northern blot analyses of the skf operon. Total RNA was isolated from wild-type strain B. subtilis 168 and a phoR null mutant. Bacteria were grown in LPM, and samples were taken 2 h before (T−2), at the time of (T0), and 5 h after (T5) entry into stationary growth phase provoked by phosphate starvation. Five micrograms of RNA was applied to each lane, and then after capillary blotting the filters were hybridized to skfA gene-specific riboprobes. (B) Northern blot analyses of the skf operon. RNA was isolated from wild-type strain B. subtilis 168. Bacteria were grown in LPM, and samples were taken 2 h before (T−2), at the time of (T0), and 2 h after (T2) entry into stationary growth phase provoked by phosphate starvation. Ten micrograms of RNA was applied per lane, and then after an alkaline-transblotting procedure the filters were hybridized to gene-specific riboprobes for skfA, skfB, and skfH. The 0.25-kb transcript was purposely run off the bottom of the gel to facilitate visualization of the weaker bands. (C) Transcriptional stability of skfA mRNA. RNA was isolated from wild-type strain B. subtilis 168, grown in LPM, and sampled 2 h after entry into stationary growth phase. RNA was isolated from samples harvested before and 2, 10, 25, 45 min after the addition of rifampin. Five micrograms of RNA was applied to each lane, and the membrane was hybridized to a skfA-specific riboprobe. Transcript sizes were determined by comparison with digoxigenin-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). An RNA molecular size ladder (0.24 to 9.5 kb) was purchased from Invitrogen.