Abstract
Haemophilus influenzae requires either heme or a porphyrin and iron source for growth. Microarray studies of H. influenzae strain Rd KW20 identified 162 iron/heme-regulated genes, representing ∼10% of the genome, with ≥1.5-fold changes in transcription in response to iron/heme availability in vitro. Eighty genes were preferentially expressed under iron/heme restriction; 82 genes were preferentially expressed under iron/heme-replete conditions.
While Haemophilus influenzae is unable to synthesize protoporphyrin IX (PPIX), it does possess a ferrochelatase that reversibly inserts iron into PPIX to form heme (13, 23). Thus, H. influenzae requires either iron, in the presence of PPIX, or heme to grow. Found only in humans, H. influenzae has evolved highly redundant mechanisms to obtain both heme and iron from host sources (16). The available heme/iron sources and the H. influenzae acquisition mechanisms have been partially characterized (6, 7, 14, 16-20, 26), although little is known about the specific regulation of these pathways.
The goals of the current study were to identify gene products potentially mediating iron and heme uptake and to provide a foundation for their further study, as well as to gain insight into possible mechanisms of regulation. Since H. influenzae can interconvert heme and iron (releasing PPIX), the genomic transcriptional profiles of growth in iron- and heme (FeHm)-replete and -restricted media were compared.
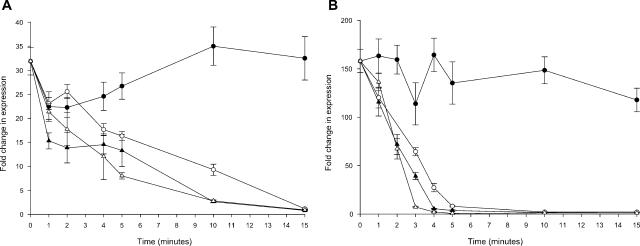
Initially the kinetics of FeHm-mediated derepression were characterized using quantitative reverse transcription-PCR (Q-RT-PCR) to examine transcripts of the FeHm-regulated iron binding protein gene hitA (1, 16). Inocula of H. influenzae Rd KW20 (ATCC 51907) were prepared as previously described (14). Inocula were added to 60-ml volumes of brain heart infusion (BHI) broth containing 10 μg/ml β-NAD (BHI-NAD) to yield ∼2 × 107 CFU/ml. FeHm-replete cultures (containing 500 μM FeCl3 and 10 μg/ml heme as hemin chloride) and FeHm-restricted cultures (containing 150 μM deferoxamine) were incubated at 37°C in a rotary shaker (175 rpm). Samples were removed at 30-min intervals for Q-RT-PCR and viable count determination as previously described (15, 27). Cells remained viable upon transfer to FeHm-restricted BHI-NAD for at least 3 h (Fig. 1A), and transcription of hitA increased to a plateau within 60 to 90 min of transfer to an FeHm-restricted environment (Fig. 1B). Conversely, transcription of hitA in FeHm-supplemented media showed no increase over the same period.
FIG. 1.
Derepression and repression kinetics defining the window of transcriptional regulation for select FeHm-regulated genes of H. influenzae Rd KW20. (A) Growth kinetics (viable count) for the FeHm-replete (closed circles) and FeHm-restricted (open circles) media. (B) Transcription of the hitA gene in FeHm-replete (closed circles) and FeHm-restricted (open circles) media. (C) Comparison of transcription of the hitA gene in medium that was FeHm replete (closed circles), FeHm restricted (open circles), or initially FeHm restricted but to which FeHm was added at 90 min (closed triangles). (D) Comparison of transcription of the genes hitA (open circles), tbp1 (open triangles), and ompP2 (closed diamonds) in medium that was initially FeHm restricted but to which FeHm was added at 90 min. The vertical dashed line indicates the point at which FeHm was added.
To determine the kinetics of FeHm repression, the experiment was repeated with supplementation of the FeHm-restricted media at 90 min with 500 μM FeCl3 and 10 μg/ml heme. Q-RT-PCR analysis demonstrated an initial increase in hitA transcripts; however, within 15 min of FeHm addition, transcripts rapidly decreased to a level identical to that in the FeHm-replete flask (Fig. 1C).
The transcription of tbp1 (11) and ompP2 (2) was also examined. These genes were considered to represent an iron/heme-regulated gene and a nonregulated gene, respectively. Genes hitA and tbp1 share a similar transcriptional profile, while ompP2 showed no changes in transcription (Fig. 1D). Oligonucleotide primers are listed in Table S1 in the supplemental data at http://peds.ouhsc.edu/lab/stull/whitby2006/tableS1.pdf.
The kinetics of Fe repression and heme repression are similar.
The above experiment was repeated with addition of iron or heme alone. There were no apparent differences after supplementation with either iron or heme alone or with both in repression of transcription of hitA or tbp2 (Fig. 2); therefore, dual-iron/heme supplementation was utilized in further studies.
FIG. 2.
Comparison of the transcription change of the genes hitA (A) and tbp2 (B) following addition of iron (open circles), heme (closed triangles), or iron and heme (FeHm) together (open triangles) or with no addition (closed circles) following growth in FeHm-restricted medium. The abscissa values are time postsupplementation of the culture medium and correspond to the region following the dotted line shown in Fig. 1C and D.
Identification of the FeHm regulon of H. influenzae.
The FeHm-restricted and FeHm-supplemented samples for microarray analysis were taken from a single flask, separated by 20 min. Three biological replicates of each condition were examined. The first sample was removed from FeHm-restricted media at 90 min postinoculation. Immediately, the flask was FeHm supplemented and a second sample was removed 20 min later. RNA for the microarray was prepared using Trizol (Invitrogen) as described by the manufacturer. Processing of the H. influenzae microarray was performed by Nimblegen (Madison, Wis.) following their protocols. Technical array replicates (arising from the presence of three arrays on each slide) were averaged prior to analysis of the biological replicates (8). The data were log2 transformed and compared between the two conditions by performing individual t tests using the TMEV software at http://www.tm4.org/mev.html (22). Genes with a 1.5-fold expression change and P < 0.05 were considered significant.
Of the 1,657 open reading frames on the Rd KW20 array, 162 were significantly differentially expressed (see Table S2 in the supplemental material at http://peds.ouhsc.edu/lab/stull/whitby2006/tableS2.pdf): 82 were maximally expressed under FeHm-replete conditions, and 80 were preferentially expressed under FeHm-restricted conditions. Comparison of microarray and Q-RT-PCR data for 11 genes indicated a linear correlation, with the microarray underestimating Q-RT-PCR values by approximately threefold.
FeHm-repressed transcripts contain genes with a known role in iron/heme uptake.
Of the 80 genes identified, 29 were single genes and 51 were in 21 putative operons. Analysis of the other genes in each operon indicated that while some genes had an increase in expression below the 1.5-fold threshold, all showed a statistically significant increase. Operons and stand-alone genes were determined based on a number of factors, including comparative genomics, transcriptional data from previous studies, conformity of orientation, size of intergenic regions, and prediction of putative transcriptional terminators using the program Transterm at http://cbcb.umb.edu/software/transterm (10). Included among the FeHm-repressible genes were known iron and heme acquisition genes (Table 1) including the hemoglobin/hemoglobin-haptoglobin receptor gene hgpC. (Strain Rd KW20 contains three potentially functional hgp genes, hgpBCD; however, both hgpB and hgpD are out of frame and not expressed.) To gain insight into possible mechanisms of regulation, we examined the upstream DNA for the presence of sequences associated with regulation by the ferric uptake regulator Fur (25). Potential sites were identified upstream of 32% of the FeHm-repressible operons.
TABLE 1.
H. influenzae Rd KW20 operons with altered transcription in response to environmental iron/heme levels
| Operon locusa | No. of genes in operonb | Gene(s)c | Fold changed
|
Fur boxe | Descriptionf | |
|---|---|---|---|---|---|---|
| Maximum | Minimum | |||||
| FeHm repressible | ||||||
| HI0017 | 1 | yfiD | −1.87 | Yes | Putative formate transferase | |
| HI0020←HI0025 | 6 | −1.75 | −1.20 | Citrate utilization operon | ||
| HI0035 | 1 | yidE | −1.55 | Putative permease | ||
| HI0075 | 1 | nrdD | −4.03 | Anaerobic ribonucleoside triphosphate reductase | ||
| HI0091←HI0092 | 2 | garK | −1.84 | −2.04 | Probable glycerate kinase and probable gluconate family permease | |
| HI0095 | 1 | −10.21 | Yes | Putative methyltransferase | ||
| HI0097→HI0099 | 3 | hitABC | −8.15 | −3.55 | Yes | ABC transporter, transport of iron across cytoplasmic membrane |
| HI0113 | 1 | −2.36 | Yes | Probable TonB-dependent protein involved in iron/heme uptake | ||
| HI0126←HI0131 | 3 | afuCBA | −1.90 | −1.62 | Yes | ABC transporter, A. pleuropneumoniae homolog involved in iron uptake |
| HI0205←HI0206 | 2 | nucA | −1.60 | −1.56 | HI0205 has no predicted function; nucA is involved in NAD uptake | |
| HI0213 | 1 | −1.68 | Yes | Putative component of ABC-type oligopeptide transport system | ||
| HI0223 | 1 | −2.45 | Yes | Predicted RarD family permease | ||
| HI0251→HI0253 | 3 | tonB, exbD, exbB | −3.02 | −2.69 | Energy transduction for heme and iron transport | |
| HI0257 | 1 | yfiA | −1.61 | Protects ribosomes during stationary-phase growth | ||
| HI0262→HI0264 | 3 | hxuCBA | −10.30 | −7.44 | Yes | Heme acquisition from hemopexin |
| HI0359←HI0362 | 4 | yfeDCBA | −11.75 | −4.35 | Yes | ABC transporter; Y. pestis homologs involved in Fe, Mn, and Zn uptake |
| HI0371←HI0380 | 10 | iscX, fdx-1, iscR | −1.88 | −1.22 | Probable involvement in Fe-S cluster formation | |
| HI0404←HI0405 | 2 | tilS, pdxY | −1.88 | −1.46 | Pyroxidine kinase (pdxY) and tRNA modification (tilS) | |
| HI0501→HI0507 | 6 | rbsGACBK | −2.50 | −1.23 | Ribose transport | |
| HI0534 | 1 | aspA | −3.12 | Conversion of aspartate to fumarate | ||
| HI0584 | 1 | −1.69 | Putative hydrolase/peptidase | |||
| HI0588 | 1 | −1.60 | Putative hydrolase/peptidase | |||
| HI0590←HI0591 | 2 | potE, speF | −3.84 | −3.20 | Uptake of putrescine (potE) and ornithine decarboxylase (speF) | |
| HI0601 | 1 | tfoX | −1.66 | Involved in regulation of competence | ||
| HI0663→HI0664 | 2 | cydDC | −1.61 | −1.61 | Yes | Probable ABC transporter |
| HI0669→HI0670 | 2 | mioC, dtd | −1.67 | −1.60 | Yes | Flavodoxin (mioC) and d-amino acid/tRNA deacylation (dtd) |
| HI0688→HI0689 | 2 | glpQ | −1.53 | −1.30 | Glycerophosphoryl diester phosphodiesterase (glpQ) | |
| HI0690→HI0691 | 2 | glpK | −1.61 | −1.44 | Glycerol uptake facilitator (glpF) and glycerol kinase (glpK) | |
| HI0712 | 1 | hgpC | −6.64 | Yes | Acquisition of heme from hemoglobin and hemoglobin-haptoglobin | |
| HI0745 | 1 | ansB | −3.42 | Periplasmic asparaginase | ||
| HI0809 | 1 | pckA | −2.87 | Phosphoenolpyruvate carboxykinase, involved in gluconeogenesis | ||
| HI0836 | 1 | genX | −1.61 | Putative tRNA synthetase | ||
| HI0863 | 1 | pdxH | −1.51 | Pyridoxamine phosphate oxidase | ||
| HI0990 | 1 | iga1 | −1.52 | Yes | Immunoglobulin A protease | |
| HI0994←HI0995 | 2 | tbp1, tbp2 | −13.94 | −12.40 | Yes | Acquisition of iron from exogenous transferrin |
| HI1189←HI1191 | 3 | −1.67 | −1.67 | Conserved hypothetical proteins | ||
| HI1210 | 1 | mdh | −2.47 | Malate dehydrogenase; role in respiration, fermentation, and gluconeogenesis | ||
| HI1217 | 1 | hup | −2.99 | Yes | Involved in uptake of heme from various sources | |
| HI1227 | 1 | uraA | −1.72 | Uracil uptake | ||
| HI1228 | 1 | upp | −1.62 | Phosphorylation of uracil to UMP | ||
| HI1275 | 1 | tehB | −3.34 | Tellurite resistance | ||
| HI1349 | 1 | −1.53 | Bacterial ferritin-like protein | |||
| HI1356→HI1360 | 5 | malQ, glgBXCA | −1.82 | −1.51 | Glycogen biosynthesis | |
| HI1368→HI1369 | 2 | pqqL | −7.02 | −1.86 | Yes | Putative protease (pqqL) and probable TonB-dependent transporter (HI1369) |
| HI1398 | 1 | fumC | −2.32 | Fumarate hydratase, reversible conversion of malate to fumarate | ||
| HI1427 | 1 | −6.12 | Putative ABC transporter, periplasmic binding protein | |||
| HI1564 | 1 | −2.04 | Probable pseudogene | |||
| HI1625←HI1626 | 2 | −1.72 | −1.38 | Conserved hypothetical proteins | ||
| HI1645 | 1 | fbp | −1.54 | Fructose-1,6-bisphosphatase, involved in gluconeogenesis | ||
| HI1660 | 1 | nrdB | −1.54 | Ribonucleoside phosphate reductase | ||
| FeHm inducible | ||||||
| HI0006→HI0009 | 4 | fdxHIE | 3.40 | 1.88 | Formate dehydrogenase-N, involved in anaerobic respiration | |
| HI0157 | 1 | fabH | 1.55 | β-Ketoacyl-acyl carrier protein synthase III, fatty acid biosynthesis | ||
| HI0164→HI0171 | 6 | nqrABCDE6 | 1.63 | 1.53 | Ubiquinone reduction and energy metabolism | |
| HI0172→HI0174 | 3 | apbE, mnmA | 1.93 | 1.69 | Putative Fe-S cluster metabolism (apbE) + tRNA (5-methyl aminomethyl-2-thiouridylate)-methyltransferase (mnmA) | |
| HI0184←HI0185 | 2 | adhC | 8.04 | 7.93 | Putative esterase (HI0184), alcohol dehydrogenase (adhC) | |
| HI0230 | 1 | nlpI | 2.30 | Conserved lipoprotein of unknown function | ||
| HI0231 | 1 | deaD | 2.48 | RNA helicase involved in rearrangement of 50S ribosome | ||
| HI0242←HI0244 | 3 | tgt | 2.30 | 1.58 | Biosynthesis of queuine in tRNA wobble positions (tgt) | |
| HI0318→HI0319 | 2 | yecO | 1.96 | 1.42 | Probable methyltransferases | |
| HI0321→HI0322 | 2 | vapB1, vapC1 | 1.64 | 1.64 | Probable toxin-antitoxin locus | |
| HI0324 | 1 | rnt | 1.63 | tRNA turnover and rRNA maturation | ||
| HI0343→HI0348 | 7 | napDGHBC | 1.93 | 1.66 | Periplasmic nitrate reductase subunits | |
| HI0381←HI0386 | 6 | tolR, ybgC | 1.52 | 1.33 | Tol-Pal system, probable transport system from outer membrane to cytoplasm | |
| HI0419 | 1 | 1.58 | Putative protease | |||
| HI0507 | 1 | 2.04 | Conserved hypothetical protein | |||
| HI0512←HI0513 | 2 | hindIIR, hindIIIM | 1.65 | 1.53 | Type 2 restriction endonuclease and methylase (HindII) | |
| HI0623→HI0625 | 3 | fmt, rrmB | 1.62 | 1.44 | tRNA modification (fmt), rRNA modification (rrmB), K+ uptake (trkA) | |
| HI0682 | 1 | ilvC | 1.55 | Ketol-acid reductoisomerase, involved in isoleucine and valine biosynthesis | ||
| HI0864 | 1 | bipA | 1.71 | Translation factor for Fis (HI0980) | ||
| HI0877←HI0878 | 2 | cgtA | 1.96 | 1.32 | Putative ribosome-associated GTPase (cgtA) | |
| HI0976→HI0978 | 3 | prmA | 1.54 | 1.29 | Methyltransferase for 50S ribosomal protein L11 (prmA) | |
| HI0979→HI0980 | 2 | dusB, fis | 2.06 | 2.00 | tRNA-dihydrouridine synthase (dusB), DNA architecture modification (fis) | |
| HI0998→HI1001 | 4 | rpL34, rnpA, yidC | 1.70 | 1.24 | Ribosomal protein (rpL34), protein component of RNase P (rnpA); hypothetical protein (HI1000), inner membrane protein translocase (yidC) | |
| HI1002 | 1 | trmE | 1.72 | GTPase involved in biosynthesis of hypermodified nucleosides in tRNA | ||
| HI1005 | 1 | 1.64 | Conserved hypothetical protein | |||
| HI1010→HI1016 | 7 | 1.86 | 1.47 | Operon contains putative transport proteins and enzymes of unknown function | ||
| HI1037 | 1 | 1.81 | Conserved hypothetical protein | |||
| HI1051 | 1 | vcaM | 1.80 | Probable multidrug resistance ABC transporter | ||
| HI1066←HI1069 | 4 | nrfABCD | 2.13 | 1.52 | Yes | Probable nitrite reductase |
| HI1075←HI1076 | 2 | cydBA | 1.72 | 1.63 | Cytochrome d ubiquinol oxidase subunits, involved in respiration | |
| HI1078←HI1080 | 3 | 1.62 | 1.22 | ABC transport system, possibly for an amino acid (by homology) | ||
| HI1089→HI1097m | 9 | ccmDEFG | 1.81 | 1.30 | Heme exporter and cytochrome c biogenesis machinery | |
| HI1173 | 1 | sprT | 1.52 | Yes | Conserved hypothetical protein | |
| HI1214→HI1216 | 3 | recJ, pfs | 1.74 | 1.43 | Single-stranded DNase (recJ); 5′-methylthioadenosine/ s-adenosylhomocysteine nucleosidase (pfs) | |
| HI1253→HI1254 | 2 | 1.76 | 1.65 | Conserved hypothetical proteins | ||
| HI1282→HI1284 | 3 | yhbC, nusA, infB | 1.96 | 1.53 | Modulator of transcription termination (nusA), essential element of translation (infB) | |
| HI1384→HI1385 | 2 | ftnA1, ftnA2 | 2.94 | 2.60 | Yes | Ferritin, iron storage proteins |
| HI1386 | 1 | 3.28 | Putative glycosyltransferase | |||
| HI1584←HI1585 | 2 | ilvHI | 2.49 | 2.36 | Acetolactate synthase III, valine biosynthesis | |
| HI1586 | 1 | 1.56 | Yes | Conserved hypothetical protein | ||
| HI1706 | 1 | betT | 3.14 | Involved in osmoprotection | ||
| HI1733 | 1 | rnb | 1.81 | RNase II, modulation of RNA decay | ||
Operon locus based on the Rd KW20 gene designation. Each arrow indicates the direction of the operon. The locus tag with the lowest number in the operon is given first for ease of reference.
Number of genes comprising the putative operon. Note that HI0006, HI0343, and HI0345 were not included on the array.
Genes contained within the operon that have been functionally characterized or identified by significant homology to other characterized genes.
Change as determined from the microarray data. Shown are the maximum and minimum values observed for genes in the putative operon.
“Yes” indicates identification of a putative Fur box based on homology to Fur consensus sequences.
Brief description of the assigned functions of gene products encoded in the operon.
Identification of other genes with a putative role in iron/heme uptake.
Several additional genes encoding proteins with putative roles in iron transport were identified, including afuABC and yfeABCD and locus tags HI0663, HI0664, and HI1427. The afu locus shares homology with an Actinobacillus pleuropneumoniae ABC iron transporter (5). While the functions of HI1427, HI0663, and HI0664 are unknown, the putative product of HI1427 has characteristics consistent with periplasmic binding proteins, while the products of HI0663 and HI0664 both have characteristics of fused permease-ATPase components of ABC transporters. The yfe locus has been shown to mediate uptake of iron, manganese, and zinc in Yersinia pestis (3). Interestingly, transcription of a putative protease gene, pqqL (HI1368), was repressed by addition of FeHm. pqqL is in an operon encoding the TonB-dependent protein HI1369. Preliminary data indicate that HI1369 has a role in heme acquisition (data not shown). In other bacterial species, proteases play a direct role in the acquisition of iron and heme from various sources (12, 21, 24, 28).
The FeHm regulon contains genes with no known role in iron/heme uptake.
A number of genes preferentially expressed under FeHm limitation appear to have a direct role on cellular metabolism. These include genes encoding the enzymes phosphoenolpyruvate carboxykinase and fructose-1,6- bisphosphatase, suggesting that there may be a cellular shift to gluconeogenesis in the absence of FeHm. Consistent with this are the observations that genes encoding aspartate ammonia lyase, asparaginase B, malate dehydrogenase, and fumerase C are repressed by FeHm addition. Together, these enzymes would lead to the conversion of l-asparagine to phosphoenolpyruvate, which then feeds into gluconeogensis to generate glucose phosphate. The latter compound may then be shunted into glycogen by the action of the products of the malQ-glg locus, also preferentially expressed under FeHm limitation.
Identification of FeHm-induced genes.
Eighty-two genes were preferentially expressed under FeHm-replete conditions: of these 17 were stand-alone genes and 65 were contained within 25 putative operons (Table 1). Several of the identified genes have a role in iron and heme processing. For example, increased transcription of the bacterial ferritin genes (ftnA1 and ftnA2) would increase the ability to store and thus detoxify excess iron. Similarly, transcription of ccmDEFG (encoding a heme exporter involved in the biosynthesis of cytochrome c) is increased. Genes having a potential role in respiration display increased transcription. These include cydAB, encoding subunits of the terminal electron acceptor cytochrome d ubiquinol oxidase, and the nqr operon encoding an Na+-translocating NADH quinone oxidoreductase. This operon is similarly upregulated in Fe-rich environments in Neisseria gonorrhoeae (9). Other operons contain genes with iron-sulfur clusters (fdxH) or heme (fdxI) as cofactors.
Q-PCR of genes not represented on the microarray.
Two operons, which were repressed by FeHm, lacked several of the predicted genes on the array. HI0007 to HI0009 were shown, by microarray, to be upregulated upon the addition of FeHm. The genes represented by these three locus tags form a putative operon with HI0006m and together encode the subunits of the nitrate-inducible formate dehydrogenase. HI0006m was considered a pseudogene due to an internal stop codon and was not included on the microarray design. However, this stop codon is conserved in other bacteria, encoding a selenocysteine residue in the E. coli HI0006m homolog (4). Comparison of RNA levels of HI0006m and HI0007 using Q-PCR confirmed that HI0006m is also repressed by addition of FeHm, consistent with the other three genes of this operon.
The transcription of HI0343 and HI0345 (both excluded from the array) was compared to levels of HI0344. This analysis determined that the genes represented by these two locus tags are also upregulated by the addition of FeHm, consistent with the other genes in that operon (data not shown).
Conclusion.
The purpose of this study was to identify H. influenzae genes affected by iron and heme uptake. Defining the regulatory kinetics characterized a minimum window most likely to represent FeHm effects. Genes known to be involved in iron and heme uptake were clearly regulated in a synchronous manner. Furthermore, significant changes in transcription of all genes in the regulated operons were demonstrated. The identification of FeHm-regulated protease and potential periplasmic transport systems provides targets for future studies. Since iron and heme are sequestered in vivo, it is likely that the pattern of regulation observed in vitro relates to the transcriptional status during infection, supporting previous studies demonstrating transcription of FeHm-repressible genes during otitis media (29).
Microarray data accession number.
Microarray data have been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE5061.
Acknowledgments
This work was supported by Public Health Service grant AI29611 from the National Institutes of Allergy and Infectious Diseases to T.L.S., D.J.M., and P.W.W. We gratefully acknowledge the support of the Children's Medical Research Institute.
We thank Jennifer Springer for technical assistance.
REFERENCES
- 1.Adhikari, P., S. D. Kirby, A. J. Nowalk, K. L. Veraldi, A. B. Schryvers, and T. A. Mietzner. 1995. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J. Biol. Chem. 270:25142-25149. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, C., E. Maier, G. Kemmer, J. Blass, A. K. Hilpert, R. Benz, and J. Reidl. 2003. Porin OmpP2 of Haemophilus influenzae shows specificity for nicotinamide-derived nucleotide substrates. J. Biol. Chem. 278:24269-24276. [DOI] [PubMed] [Google Scholar]
- 3.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 4.Berg, B. L., J. Li, J. Heider, and V. Stewart. 1991. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J. Biol. Chem. 266:22380-22385. [PubMed] [Google Scholar]
- 5.Chin, N., J. Frey, C. F. Chang, and Y. F. Chang. 1996. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 143:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Cope, L. D., R. P. Love, S. E. Guinn, A. Gilep, S. Usanov, R. W. Estabrook, Z. Hrkal, and E. J. Hansen. 2001. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect. Immun. 69:2353-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, X., and G. A. Churchill. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4:210. [Online.] doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 11.Gray-Owen, S. D., and A. B. Schryvers. 1995. Characterization of transferrin binding proteins 1 and 2 in invasive type b and nontypeable strains of Haemophilus influenzae. Infect. Immun. 63:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, X., A. Sroka, J. Potempa, and C. A. Genco. 2004. Coordinate expression of the Porphyromonas gingivalis lysine-specific gingipain proteinase, Kgp, arginine-specific gingipain proteinase, RgpA, and the heme/hemoglobin receptor, HmuR. Biol. Chem. 385:1049-1057. [DOI] [PubMed] [Google Scholar]
- 13.Loeb, M. R. 1995. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J. Bacteriol. 177:3613-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morton, D. J., L. L. Madore, A. Smith, T. M. VanWagoner, T. W. Seale, P. W. Whitby, and T. L. Stull. 2005. The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol. Lett. 253:193-199. [DOI] [PubMed] [Google Scholar]
- 15.Morton, D. J., A. Smith, L. L. Madore, T. M. VanWagoner, T. W. Seale, P. W. Whitby, and T. L. Stull. 2004. Identification of a haem utilization protein (Hup) in Haemophilus influenzae. Microbiology 150:3923-3933. [DOI] [PubMed] [Google Scholar]
- 16.Morton, D. J., and T. L. Stull. 2004. Haemophilus, p. 273-292. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. American Society for Microbiology, Washington, D.C.
- 17.Morton, D. J., T. M. VanWagoner, T. W. Seale, P. W. Whitby, and T. L. Stull. 2006. Utilization of myoglobin as a heme source by Haemophilus influenzae requires binding of myoglobin to haptoglobin. FEMS Microbiol. Lett. 258:235-240. [DOI] [PubMed] [Google Scholar]
- 18.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morton, D. J., and P. Williams. 1989. Utilization of transferrin-bound iron by Haemophilus species of human and porcine origins. FEMS Microbiol. Lett. 53:123-127. [DOI] [PubMed] [Google Scholar]
- 20.Morton, D. J., and P. Williams. 1990. Siderophore-independent acquisition of transferrin-bound iron by Haemophilus influenzae type b. J. Gen. Microbiol. 136:927-933. [DOI] [PubMed] [Google Scholar]
- 21.Otto, B. R., S. J. van Dooren, C. M. Dozois, J. Luirink, and B. Oudega. 2002. Escherichia coli hemoglobin protease autotransporter contributes to synergistic abscess formation and heme-dependent growth of Bacteroides fragilis. Infect. Immun. 70:5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 23.Schlör, S., M. Herbert, M. Rodenburg, J. Blass, and J. Reidl. 2000. Characterization of ferrochelatase (hemH) mutations in Haemophilus influenzae. Infect. Immun. 68:3007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigematsu, T., J. Fukushima, M. Oyama, M. Tsuda, S. Kawamoto, and K. Okuda. 2001. Iron-mediated regulation of alkaline proteinase production in Pseudomonas aeruginosa. Microbiol. Immunol. 45:579-590. [DOI] [PubMed] [Google Scholar]
- 25.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 26.Stull, T. L. 1987. Protein sources of heme for Haemophilus influenzae. Infect. Immun. 55:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanWagoner, T. M., P. W. Whitby, D. J. Morton, T. W. Seale, and T. L. Stull. 2004. Characterization of three new competence-regulated operons in Haemophilus influenzae. J. Bacteriol. 186:6409-6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitby, P. W., T. M. VanWagoner, J. M. Springer, D. J. Morton, T. W. Seale, and T. L. Stull. 2006. Burkholderia cenocepacia utilizes ferritin as an iron source. J. Med. Microbiol. 55:661-668. [DOI] [PubMed] [Google Scholar]
- 29.Whitby, P. W., K. E. Sim, D. J. Morton, J. A. Patel, and T. L. Stull. 1997. Transcription of genes encoding iron and heme acquisition proteins of Haemophilus influenzae during acute otitis media. Infect. Immun. 65:4696-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]