Abstract
The broad-host-range plasmid RK2 is capable of replication and stable maintenance within a wide range of gram-negative bacterial hosts. It encodes the essential replication initiation protein TrfA, which binds to the host initiation protein, DnaA, at the plasmid origin of replication (oriV). There are two versions of the TrfA protein, 44 and 33 kDa, resulting from alternate in-frame translational starts. We have shown that the smaller protein, TrfA-33, and its 64-residue amino-terminal peptide (designated T1) physically interact with the Escherichia coli β sliding clamp (β2). This interaction appears to be mediated through a QLSLF peptide motif located near the amino-terminal end of TrfA-33 and T1, which is identical to the previously described eubacterial clamp-binding consensus motif. T1 forms a stable complex with β2 and was found to inhibit plasmid RK2 replication in vitro. This specific interaction between T1 and β2 and the ability of T1 to block DNA replication have implications for the previously reported cell lethality caused by overproduction of T1 (P. D. Kim, T. M. Rosche, and W. Firshein, Plasmid 43:214-222, 2000). The toxicity of T1 was suppressed when wild-type T1 was replaced with mutant T1, carrying an LF deletion in the β-binding motif. Previously, T1 toxicity has been shown to be suppressed by Hda, an intermediate regulatory protein which helps prevent overinitiation in E. coli through its interaction with the initiator protein, DnaA, and β2. Our results support a model in which T1 toxicity is caused by T1 binding to β2, especially when T1 is overexpressed, preventing β2 from interacting with host replication proteins such as Hda during the early events of chromosome replication.
In Escherichia coli, the sliding clamp β2 subunit of the DNA polymerase III holoenzyme is a central component of DNA replication machinery, playing a pivotal role in the DNA replication cycle both during replication and in the switching from initiation to elongation. This dimeric ring protein confers processivity to the core polymerase (αɛθ) by binding directly to the catalytic α subunit of the core and tethering it to DNA during replication (20, 35). β2 is loaded onto DNA by the single δ subunit in the clamp loader (γ complex) which interacts with β2 to open its subunit interfaces and allow its assembly onto double-stranded DNA (39). β2 also plays a role in DNA repair, as several other polymerases, including polymerase II (Pol II, or PolB) and Pol IV (or DinB), were shown to interact with β2 (3, 4, 25, 40), and in the mutant β2 strain (dnaN159), DNA replication, repair, and translesion synthesis are altered (37). Pol V (or UmuD/C), which is capable of error-prone replication past sites of DNA damage (translesion synthesis), also binds to β2 (38). In addition to the repair polymerases, DNA polymerase I, MutS (a mismatch recognition protein), and DNA ligase have also been shown to physically interact with β2 (26). β2 may, in addition, be involved in the early events of chromosome replication (11, 24, 36) via DnaA, the replication initiator protein which recognizes the origin of replication (oriC) to initiate replication (33). The interaction between DnaA, β2 (complexed with DNA), and the recently identified host initiation protein, Hda (14), helps prevent unprogrammed initiation of additional cycles of replication. The DnaA-Hda-β2 complex helps convert the active ATP-bound form of DnaA (which is required for reinitiation) to an inactive ADP-bound form through accelerated ATP hydrolysis (12, 13, 24, 36).
The utilization of the versatile β2 sliding clamp by so many polymerases, as well as its interaction with the δ subunit of the γ complex (for an example, see reference 35), Hda (24), and other proteins raises questions about the positions and interplay of binding sites on the β2 ring. For example, the α and δ subunits of Pol III holoenzyme are thought to have overlapping interaction sites on the β2 molecule (29). For many proteins, the interaction with β2 is mediated by a pentapeptide motif with consensus sequence QL[D/S]LF or a related hexapeptide with the consensus sequence QL[SP]LPL (7, 8, 41). These motifs are required for several polymerases and replication accessory proteins, including DnaE (α), PolC, PolB, DinB, UmuC, and Hda, to bind specifically to E. coli β2 by making contact with a hydrophobic pocket located at the base of the C-terminal tail of each β monomer. Based on the identification of the pentapeptide motif, a conserved SLF motif in the δ subunit was proposed to be involved in the binding of δ to β2 during loading of β2 onto DNA (7). The crystallographic structures of the β2-δ complex (10) and of the Pol IV (DinB) fragments complexed with β2 (3, 4) confirmed that LF residues on the β2-binding motif of E. coli δ and LL residues on DinB make most of the intermolecular contact and penetrate a hydrophobic pocket on the β2 surface. Since β2 interacts with various replication proteins, both essential and accessory, it is expected that proteins or molecules that can bind to β2 with high potency will inhibit DNA replication and result in cell death.
The broad-host-range plasmid RK2 encodes the essential replication initiation protein, TrfA, which binds to the host initiation protein, DnaA, at the plasmid origin of replication (oriV) to form an open complex in oriV (22). Two forms of the TrfA protein, 44 kDa (TrfA-44) and 33 kDa (TrfA-33), generated by two in-frame translational start sites spaced 97 amino acids apart, are encoded by RK2 (34). A toxic peptide (designated T1) derived from the amino-terminal portion of TrfA (residues 99 to 163 from TrfA-44 coordinates) (34) has been identified (16). This toxic phenotype can be suppressed by the overexpression of Hda (17). As Hda functions in preventing overinitiation in E. coli, the observation suggested that the toxicity of the T1 peptide may be due to interference with the activity of Hda or other host proteins involved in DNA replication initiation. Furthermore, this interference is related to the altered structure or conformation of T1 (relative to the full-length protein), as TrfA proteins and other peptides derived from TrfA, including the longer version of T1 (T1-2), are not toxic to E. coli (16).
In this study, we show for the first time that the smaller form of the broad-host-range plasmid RK2 Rep protein, TrfA-33, and its peptide T1 both can interact with β2 and that the interaction is mediated through the QLSLF sequence of TrfA-33 protein and T1 peptide. Using an in vitro replication system for plasmid RK2, we have shown that T1 can inhibit DNA replication. Furthermore, we have demonstrated that the toxicity of T1 was totally suppressed by replacing wild-type T1 with mutant T1 which lacks the LF in the QLSLF β2-binding motif. The double mutation abolishes not only the ability of T1 to bind to β2 but also T1's inhibitory effect on RK2 replication in vitro. Taken together, these results support the physiological relevance of the protein-protein interactions involving T1 and β2 during chromosome replication initiation and cell lethality.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
XL1-Blue from Stratagene was used for subcloning, TOP10 and BL21(DE3)/pLyS cells (Invitrogen) were used to overexpress various TrfA-33 peptide constructs and for the production of TrfA-33 proteins, and C600 was used for the preparation of cell extracts. Ampicillin, isopropyl-β-d-thiogalactopyranoside (IPTG) and l-(+)-arabinose were purchased from Sigma, and [methyl-3H]dTTP was purchased from ICN Radiochemicals. The penta-His antibody for immunoblot assays was from QIAGEN. The restriction enzymes were obtained from New England Biolabs and Promega and the ligation kit from Roche. Oligonucleotides for PCR were purchased from Sigma Genosys. Bacteria were grown in Luria-Bertani (LB) broth (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl/liter) or terrific broth (12 g of tryptone, 24 g of yeast extract, and 4 ml of glycerol/liter) with 100 μg/ml of ampicillin. The plasmids used in this study include pBAD vector (Invitrogen) that bears the N-terminal six histidine residues to allow the expression of N-terminal His-tagged polypeptide fragments after induction with l-(+)-arabinose from the araBAD promoter, the T7 expression vector pET-16b from Novagen (IPTG inducible) that provides the amino-terminal His tag, and pGEM-T vector (Promega) for subcloning of PCR products. Peptides containing the β2-binding pentamer motif (pep14) and a related control peptide (pep4) were synthesized on a PerSeptive Pioneer Peptide Synthesis System as previously described (7). Crude peptides were purified by high-performance liquid chromatography, and their concentrations were estimated by analytical reverse-phase high-performance liquid chromatography, amino acid analysis, and mass spectrometry.
Cloning and expression of His6-tagged TrfA peptides.
Portions of the trfA gene were amplified by PCR using primers essentially as described by Kim et al. (16), with plasmid RK2 as the template in the reaction mixtures. The primers were designed based on those described by Kim et al. (16), with NcoI and HindIII sites added to the 5′ and the 3′ ends, respectively, of each trfA fragment for cloning purposes. These fragments, which encode amino acids 99 to 163 for T1, amino acids 164 to 234 for T2, and amino acids 99 to 234 for T1-2 portions of TrfA-33, were cloned into pBAD/MycHisB vector to yield the pBAD-T1, pBAD-T2, and pBAD-(T1-2) constructs, respectively.
Site-directed mutagenesis.
The gene fragments encoding the full-length TrfA-33 and T1 peptide (amino acids 99 to 163 of the TrfA protein) were initially cloned into pGEM-T vector (Promega). The trfA-33 gene was synthesized by PCR from pRK2 plasmid using the primers 5′-GTATCCATGGCGACCAAGAAGCGAAAAACCGCC-3′ and 5′-CCAGCTCGAGGCGTTTGCAATGCACCAGGTC-3′ (the restriction enzyme NcoI and HindIII sites are underlined). These plasmids were used as templates for the generation of LF deletion mutants using the QuikChange site-directed mutagenesis kit (Stratagene). The primers used for site-directed mutagenesis were 5′-GAAATGCAGCTTTCCGATATTGCGCCGTGG-3′ and CCACGGCGCAATATCGGAAAGCTGCATTTC-3′. The mutant clones were sequenced using the Big Dye Terminator ready reaction sequencing kit (Perkin Elmer Life Sciences) to confirm the presence of the deletion mutation and the authenticity of the remaining sequence. To generate TrfA-33 (ΔLF) and T1 (ΔLF) expression plasmids, the XhoI fragment of TrfA-33 and the NcoI-HindIII fragment encoding mutant T1 peptide were cloned into pET16b and pBAD vectors, respectively.
TrfA-33 and β2 proteins expression and purification.
His-tagged wild-type and mutant forms of TrfA-33 (see below) proteins were purified from BL21(DE3)/pLysS cells transformed with the pET-16b-TrfA-33 and pET-16b-TrfA-33 (ΔLF) plasmids. Purification was undertaken according to the standard method described by the manufacturer (Novagen). Briefly, lysates were prepared from IPTG (0.5 mM)-induced cells by repeated freeze-thaw, followed by sonication in buffer containing 50 mM Tris (pH 7.4), 500 mM NaCl, 0.1% Triton X-100, 10% glycerol, 10 mM β-mercaptoethanol, 10 mM imidazole, and 0.5 mg/ml lysozyme. After lysis, clarified supernatants were loaded onto a Ni2+-nitrilotriacetic acid (NTA) agarose column equilibrated with the above buffer with lysozyme omitted. TrfA-33 proteins were eluted with gradients of imidazole in elution buffer (50 mM Tris [pH 7.4], 500 mM NaCl, 10% glycerol, 0.1 mM dithiothreitol). Fractions containing proteins, identified by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were pooled and dialysed against 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol (storage buffer) at 4°C. All preparations yielded samples with greater than 85% purity as judged by Coomassie staining.
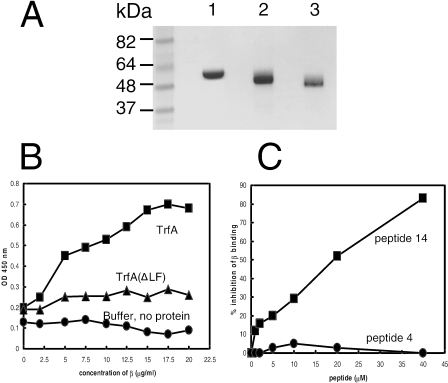
The E. coli β2 protein was prepared as described by Oakley et al. (30). Fractions containing β2, identified by Coomassie-stained SDS-PAGE, were pooled and dialyzed against storage buffer. The preparation yielded samples greater than 95% pure, as judged by scanning densitometry of Coomassie-stained SDS-PAGE (see Fig. 2A).
FIG. 2.
(A) SDS-PAGE analysis of purified wild-type β2 (lane 1), His-tagged TrfA-33 (lane 2), and His-tagged TrfA-33 (ΔLF) (lane 3) proteins. Three to five micrograms of each purified protein was electrophoresed through a 4 to 12% SDS-PAGE gel prior to being stained with Coomassie brilliant blue R-250 (Bio-Rad). Positions of size standards are indicated. (B) Interactions of β2 with TrfA-33 proteins. Increasing concentrations of β2 protein were added to TrfA-33 proteins immobilized onto microtiter plates (5 μg/ml, 50 μl/well). Bound β2 proteins were detected with anti-β2 antibody. ▪, ΤrfA-33 on plate; ▴, TrfA-33 (ΔLF) on plate; •, binding buffer, no protein on plate. (C) β2-binding consensus peptide, peptide 14 with the sequence IG QLSLF GV, inhibited the interaction of TrfA-33 and β2. Microtiter plate assay results showing the inhibition of TrfA-33 and β2 interaction by addition of various amounts of peptide 14. No inhibition was observed with the control peptide, peptide 4 with the sequence IG QADMA GV. ▪, peptide 14; •, peptide 4.
Small-scale purification of His-tagged TrfA peptides under native conditions.
Six-His-tagged T1, T1 (ΔLF), T2, and T1-2 peptides were expressed from pBADMycHisB plasmids and purified as described by QIAGEN (QIAexpress Ni-NTA Fast Start Handbook) from 250-ml to 1-liter cultures of E. coli TOP10 bearing pBAD plasmids overproducing various TrfA peptides. Cultures were grown in LB medium containing ampicillin (100 μg/ml) at 30°C. When cultures reached an optical density at 600 nm (OD600) of ∼0.8, arabinose was added to a final concentration of 0.02 or 2% (0.02% for T1 and 2% for other constructs) and growth continued for an additional 3 h. Cells were harvested by centrifugation, resuspended in 10 ml of 25 mM Tris-HCl (pH 8.0), and lysed in the lysis buffer supplemented with lysozyme and Benzonase (QIAGEN). After centrifugation at 16,000 rpm for 30 min at 4°C, the soluble fraction was applied to a Ni2+-NTA column, and bound His-tagged peptides were eluted with two 1-ml aliquots of elution buffer. Washed and eluted fractions were then analyzed by SDS-PAGE.
Microtiter plate binding assay/binding inhibition assays.
Plate binding was performed as described previously (7), and assays were done in triplicate. Briefly, purified TrfA-33 was diluted in coating buffer (5 μg/ml) and immobilized onto 96-well microtiter plates (Falcon flexible plates; BD) by overnight incubation (50 μl/well) at 4°C. Subsequent steps were done at room temperature. Plates were blocked for 1 h in 5% nonfat dry milk in WB3 (100 μl/well; 20 mM Tris, pH 7.5, 0.1 mM EDTA containing 0.05% [vol/vol] Tween 20) at room temperature, washed, and incubated for 1 h with 0 to 20 μg/ml β2 in binding buffer (10 mM Tris, pH 7.5, 10 mM MgCl2, 0.1 mM EDTA). After binding, plates were incubated with rabbit anti-β2 (1:1,000, 50 μl/well, 1 h), followed by incubation with goat anti-rabbit conjugated to horseradish peroxidase (1:1,000, 50 μl/well, 1 h). Color development was achieved by 2′,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid diammonium salt (ABTS)-horseradish peroxidase reaction. Absorbance was read at A405 using a Multiskan reader with Ascent software (Labsystems).
For the peptide inhibition assay, peptides (the β2-binding consensus pep14, IG QLSLF GV, and the control peptide, pep4 with sequence IG QADMA GV) were allowed to complex with β2 (10 μg/ml in binding buffer, as described above) in a preblocked 96-well microtiter plate (Sarstedt) for 90 min at room temperature. Aliquots of 50 μl were then transferred from each well to a corresponding well of the TrfA-33-coated plates. After 15 to 20 min of incubation, the amount of bound β2 was detected as described above.
Ni-NTA pull down of β2.
Taking advantage of the histidine-tagged amino terminus of TrfA-33, T1, T2, and T1 (ΔLF), Ni-NTA pull down of β2 was carried out as described previously (24). Eluates were separated by SDS-PAGE, followed by immunoblotting with anti-β2 antibody. β2 bands were visualized by chemiluminescence (Pierce).
Effects of various TrfA peptide overproduction on the viability of E. coli cells.
The effect of TrfA peptide (T1, T2, T1-2, and T1 mutant) overproduction during growth was investigated in liquid medium. E. coli TOP10 strains transformed with pBAD-T1, pBAD-T2, pBAD-(T1-2), and pBAD-T1 (ΔLF) were grown overnight in LB medium with 100 μg/ml ampicillin at 37°C. The culture was diluted 1:50 in the same medium with arabinose (2%) to induce peptide expression. The induced cultures were incubated at 37°C, and at various time points, samples were withdrawn and their OD600 measured.
Measurement of chromosome DNA synthesis.
All DNA synthesis assays were performed in duplicate. Briefly, overnight cultures of TOP10 cells transformed with TrfA peptide constructs were diluted 1:50 in LB medium containing 0.1 μM [methyl-3H]dTTP, ampicillin (100 μg/ml), and 2% arabinose and then grown at 37°C. During growth, aliquots of cells (200 μl) were taken at various times and treated with ice-cold 10% trichloroacetic acid. Acid-insoluble fractions were collected on GF/C filters (Whatman), and radioactivity was measured using a liquid scintillation counter.
Plasmid RK2 replication using crude extracts.
Preparation of the E. coli crude fraction II extract and reaction conditions were carried out essentially as described previously (19). Standard reaction mixtures contained 300 ng of supercoiled plasmid RK2 DNA template; 40 mM HEPES, pH 8; 50 μg/ml bovine serum albumin; 10 mM MgCl2; 2 mM ATP; 500 μM (each) GTP, CTP, and UTP; 50 μM of each deoxynucleoside triphosphate; 150 pmol/ml [methyl-3H]dTTP; 80 mM creatine phosphate; 5% polyethylene glycol; 50 mM KCl; 16 μg/ml creatine kinase; and 250 ng of the histidine-tagged version of TrfA-33 protein. Reactions were carried out at 30°C for 2 h and were stopped by adding 0.1 M sodium pyrophosphate, and incorporated counts were determined after precipitation with 10% trichloroacetic acid. The RK2 plasmid was purified by using the S.N.A.P. MidiPrep kit (Invitrogen).
RESULTS
Identification of a putative β2-binding motif in TrfA and related plasmid-borne DNA replication (Rep) proteins.
The amino acid sequence of TrfA was used in a series of PSI-BLAST searches of the nonredundant protein database at GenBank. Two families of protein were identified in this search: those closely related in sequence to TrfA and a larger more diverse family of plasmid-borne replication proteins related to plasmid pMLb RepA. A nonredundant set of proteins was generated, and the protein sequences were aligned using MAFFT (v5.662) (15). A single conserved putative β2-binding motif related to the pentapeptide QL[SD]LF was identified close to the amino terminus of the short form of TrfA (Fig. 1).
FIG. 1.
Alignment of partial amino acid sequences of TrfA/RepA orthologues from plasmids related to RK2 and pMLb. Amino acids matching the beta-binding site consensus sequence are underlined. The vertical arrow indicates the start of the TrfA-33 protein. Sequences are labeled with the plasmid/mobile genetic element of origin or GenBank gi number where the plasmid/mobile genetic element has not been named.
TrfA-33 can bind β2, and the binding is inhibited by the peptide IGQLSLFGV.
To determine that TrfA-33 interacts with β2, we used plate assays with purified recombinant E. coli His10TrfA-33 (TrfA-33 with a 10-histidine tag at the amino terminus), purified wild-type β2 (Fig. 2A), and polyclonal antibody to the β2 polypeptide. Τhe binding curves of β2 to immobilized TrfA-33 on plates (Fig. 2B) showed that the amount of β2 bound to TrfA-33 increased with increasing concentrations and saturated at higher levels. Our previous work showed that peptides containing the consensus pentapeptide motif QL[SD]LF can inhibit α-β2, δ-β2, and Hda-β2 interactions in vitro (7, 24, 41). These pentapeptides can also inhibit in vitro DNA synthesis (41). If the interaction of β2 with TrfA-33 is indeed mediated through the pentapeptide sequence of TrfA-33, one would expect that the binding of TrfA-33 to β2 would be disrupted by the addition of the consensus peptide (peptide 14), which has the sequence IG QLSLF GV. That this is the case is shown in Fig. 2C. The addition of 40 μM peptide 14 resulted in about 85% inhibition of binding of TrfA-33 to β2. A 50% inhibition of binding was achieved at ∼20 μM of the peptide. No inhibition of TrfA-33-β2 interaction was observed when the control peptide, peptide 4 (IG QADMA GV), was used (Fig. 2C).
To confirm the location of the beta binding motif, a mutant of TrfA-33 which was defective in the putative β2-binding site identified by the bioinformatics analysis (Fig. 1) was constructed. This mutant would be predicted to have severe disruption in its ability to bind β2, and that was the case. His10TrfA-33 (ΔLF) exhibited severely diminished affinity for β2 in the plate assay (Fig. 2B). Taken together, these results demonstrate that the QLSLF motif close to the amino terminus of TrfA-33 mediates binding of TrfA-33 to β2.
A peptide derived from the amino-terminal portion of TrfA-33 containing the putative beta binding motif (T1) is toxic to host cells.
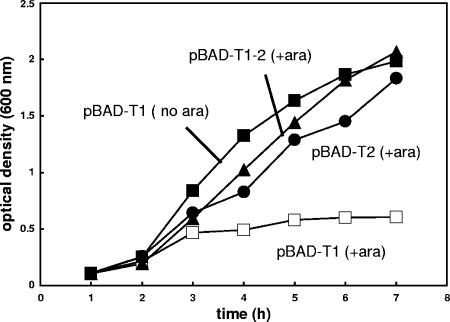
Kim et al. (16) recently reported that the fragment of TrfA-33 (T1 peptide) containing amino acids 99 to 163 (TrfA-44 coordinates) could not be expressed because it appeared to be toxic to the host strain. The same authors also demonstrated that T1 toxicity was partially eliminated when the longer polypeptide T1-2, consisting of the amino acids 99 to 234 region of TrfA-33, was expressed. To confirm the toxicity of T1, we used E. coli strain TOP10 transformed with various pBAD plasmids harboring TrfA-33 fragments, pBAD-T1, pBAD-T2, and pBAD-(T1-2), all regulated by the araBAD promoter. Our results confirmed those of Kim et al. (16) and demonstrated that overproduction of T1 was toxic to host cells, but no inhibitory effect on growth was observed with overexpression of either T2 or T1-2 (Fig. 3). The toxicity was dependent on the amount of T1 produced in the cells, as cell growth was not affected without T1 induction and the pBAD system can express both soluble T2 and T1-2 peptides at a very high level following arabinose induction (data not shown). We also examined DNA replication and cell morphology of E. coli cells overproducing T1 and found that both cell growth (Fig. 3A) and DNA synthesis (Fig. 3B) were severely suppressed and the cells expressing T1 peptide exhibited an extremely elongated morphology with no clear constrictions (Fig. 4, left panel). This filamentous phenotype is consistent with those observed in cultures of E. coli after inhibition of initiation of chromosome replication (23). Taken together, these results suggest that T1 interferes with essential DNA replication and cell cycle control in E. coli. The fact that T1 peptide includes the beta binding site further suggests that β2-binding may be responsible for the toxic effect of the peptide.
FIG. 3.
Inhibition of cell growth upon overproduction of T1. E. coli TOP10 cells were transformed by various pBAD plasmids expressing TrfA-33 peptides. Cells were grown at 37°C, and protein expression was induced by adding arabinose (final concentration, 2%) to the culture. Aliquots were then withdrawn at the indicated times for measurement of the optical density (OD600) which was used as an indicator of cell growth. □, pBAD-T1 with 2% arabinose added; ▪, pBAD-T1 with no arabinose; ▴, pBAD-T1-2 with 2% arabinose added; •, pBAD-T2 with 2% arabinose added.
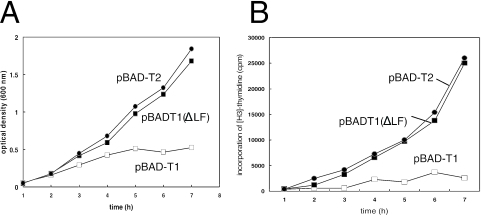
FIG. 4.
Growth, DNA replication, and cell division were suppressed by overproduction of the TrfA T1 peptide but not by the TrfA T1 (ΔLF) mutant. Cells were transformed by various pBAD plasmids and grown at 37°C in LB supplemented with 100 μg/ml ampicillin and 2% arabinose. Cultures were withdrawn at the times indicated. (A) Cell growth was monitored by measuring the OD600 of the cultures. (B) DNA replication was assessed by measuring the incorporation of [H3]dTTP into acid-insoluble materials. □, pBAD-T1; ▪, pBAD-T1 (ΔLF); •, pBAD-T2.
Deletion mutant in the β2-binding motif of T1 can suppress the inhibitory effect of T1 overexpression.
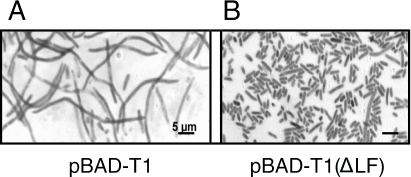
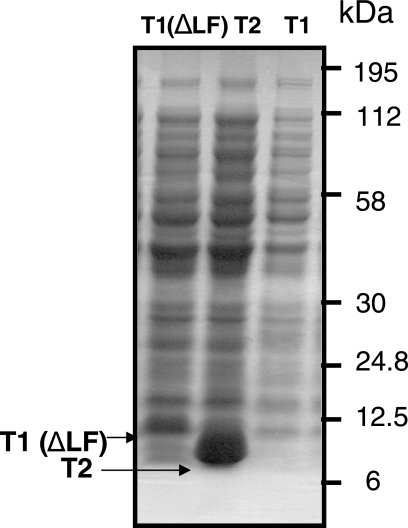
Next we studied the effect of deletion of the LF residues from the β2-binding motif of T1. We studied the ability of the deletion mutant to reverse the inhibitory effect of T1 on cell growth as well as DNA synthesis and the filamentous phenotype. As in our experiments described above, we used the TOP10 strains transformed with pBAD-T1, pBAD-T1 (ΔLF), and pBAD-T2 (as a control). We found that, contrary to the wild type, mutant T1 peptide [T1 (ΔLF)] overproduction had no effect on cell growth and DNA synthesis (Fig. 3A and B). Introduction of pBAD-T1 (ΔLF) into TOP10 cells could also complement cell cycle defects associated with wild-type T1 (Fig. 3 and 4). Not only did most of the cells overexpressing T1 (ΔLF) appear capable of completing cell division they also accumulated the mutant peptide at substantial amounts within 3 h of arabinose induction (Fig. 5B). These results indicate that the β2-binding site of T1 is responsible for the toxic phenotype of the wild-type peptide.
FIG. 5.
Defects in cell morphology in the cells with overexpressed T1 peptide and suppression of this phenotype in the cells expressing the mutant peptide. Cells transformed with pBAD-T1 and pBAD-T1 (ΔLF) were grown at 37°C in LB supplemented with 100 μg/ml ampicillin and 2% arabinose until the OD600 reached 0.5. They were then fixed with methanol and stained with neutral red (0.5%) to obtain microphotographs. Bars, 5 μm.
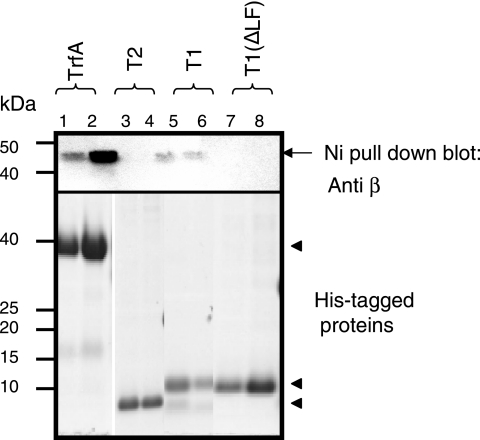
LF in the QLDLF motif of T1 is critical for T1-β2 interaction.
Next, we looked at whether the LF residues are required for T1 for the interaction with β2. His-tagged versions of TrfA-33, T1, T2, and T1 (ΔLF) were allowed to bind to β2, and the protein was pulled down with Ni-NTA agarose beads. Immunoblotting of the pull down for the presence of β2 reveals the ability of the protein to stably bind β2. Using this assay, wild-type TrfA-33 (Fig. 6, lanes 1 and 2) and wild-type T1 (Fig. 6, lanes 5 and 6) effectively interacted with β2; however, neither T2 (Fig. 6, lanes 3 and 4) nor the double mutant T1 (Fig. 6, lanes 7 and 8) could be detected in the pull down, indicating that the LF deletion mutant of T1 is unable to form a stable complex with β2.
FIG. 6.
T1 interacts with β2, and mutation in β2-binding motif disrupts the interaction. Purified H10-TrfA-33, H6-T1, H6-T2, and H6-T1 (ΔLF) were coupled to 50 μl of Ni-NTA resin (2 h at 4°C with rotation) and incubated with 2 μg of wild-type β2 in 200 μl of binding buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, and 0.05% Tween 20) at room temperature for 30 min with constant agitation. After excessive washing with the same buffer (four times with 500 μl), the resin was resuspended in 25 μl of SDS sample buffer, and the bound proteins were separated in SDS-PAGE and probed with anti-β2 polyclonal antibody. Arrows indicate histidine-tagged proteins, and the arrow indicates the Ni-NTA pull-down product, β2.
T1, but not T1 (ΔLF), inhibits RK2 replication in vitro.
T1 was also analyzed for its effect on DNA replication in vitro using an assay based on soluble E. coli extract (fraction II) to replicate supercoiled DNA containing an RK2 origin (oriV) in the presence of the initiator protein TrfA (19). When T1 was added to the reaction mixture, the in vitro replication activity was substantially reduced to approximately 6% of the control reaction. In contrast, the addition of T2 or T1 (ΔLF) peptide had little effect on the replication activity (Table 1).
TABLE 1.
T1 peptide inhibition of RK2 replicationa
| Peptide (amt [μg]) added to replication assay | Amt of DNA (pmol) | % of DNA |
|---|---|---|
| None | 167 | 100 |
| T1 (0.5) | 10 | 5.9 |
| T1 (ΔLF) (0.5) | 155 | 92.8 |
| T2 (0.5) | 132 | 79 |
In vitro replication was performed using an E. coli C600 extract (fraction II) active for RK2 replication. Reaction mixtures contained 300 ng of supercoiled RK2 plasmid and 250 ng of His10TrfA-33. Total nucleotide incorporation and relative replication activity are shown.
DISCUSSION
Many plasmids code for their own replication initiation proteins (Rep); for example, the broad-host-range plasmid RK2 synthesizes two variants of the TrfA initiation protein (33-kDa and 44-kDa forms). RK2 plasmid replication requires the participation of one or other of these TrfA initiating proteins, depending on the host (9, 21). Genetic analyses suggest that the interactions between Rep proteins and several host replication proteins are required for the normal replication of the plasmid. In the case of the broad-host-range plasmid RK2, initiation of DNA replication from the plasmid origin oriV requires the binding of either form of TrfA protein to the host DnaA (22). This binding results in the formation of an open complex or “replication bubble” at oriV as well as serving to recruit and position the DnaB replicative helicase at this open region (21, 22). Recently, TrfA has also been found to interact with Hda (for “homologous to DnaA”), the AAA+ E. coli protein which is required for inhibition of DNA synthesis initiation via antagonism of DnaA activity by β2 (24, 36). Overexpression of Hda was shown to inhibit both maintenance and replication of RK2 (1). In this study, we have demonstrated that TrfA-33 physically interacts with β2 and that this interaction is required by RK2 for its replication, as His10TrfA-33 (ΔLF) did not support the RK2 replication in vitro (data not shown). It is evident that TrfA proteins play an essential role in RK2 replication initiation and that they do this through their interactions with several host proteins, including recruitment of DnaA, DnaB-DnaC complex, Hda, and β2 to the origin of replication of the plasmid RK2. The amino alignments (Fig. 1) predict that most, if not all, of the RepA orthologues encoded by plasmids related to pMLb will also recruit β2 to their origins of replication.
Kim et al. (16, 17) showed that expression of the membrane-binding fragment of TrfA (T1), encoding amino acids 99 to 163, was lethal in E. coli, and Hda was identified as a suppressor of this lethality (17). This lethality does not involve replication of the RK2 plasmid, only chromosome replication in which TrfA presumably plays no part. We have confirmed that overexpression of the T1 fragment of TrfA is lethal and have further demonstrated that the inhibition of cell growth is most likely to result from the inhibition of host chromosomal DNA replication and cell division blockage (Fig. 3 and 4). Further evidence that T1 interferes with DNA replication came from the inhibitory effect of T1 on the plasmid RK2 replication in an in vitro replication system (Table 1). Furthermore, our experiments showed that T1 could form a stable complex with β2 (Fig. 6). Mutating the β2-binding motif of T1 not only disrupted the T1-β2 protein-protein interaction (Fig. 6) but also suppressed the cellular toxicity of T1 (Fig. 4 and 7) and abolished T1's inhibitory effect on plasmid RK2 replication (Table 1). Taken together, the results strongly suggest that the molecular basis of the T1-induced host DNA replication inhibition and the subsequent cellular toxicity is the blockage of the QL[S/D]LF-binding site of the β2 protein by the overproduced T1.
FIG. 7.
T1 (ΔLF) can be expressed at a high level in E. coli cells, but T1 cannot. Coomassie-stained SDS-PAGE of total cell lysates from TOP10 cells expressing T1 (ΔLF), T2, and T1. Arrows point to His-tagged T1 (ΔLF) and T2. Protein size standards are shown on right.
It should be noted, however, that the lack of affinity of T1 (ΔLF) for β2 (Fig. 6) and the observed lack of toxicity (compared to the “native” T1 peptide) (Fig. 4 and 7) may not be due to the absence of the essential LF residues but rather to a “misfolding” of another motif or the incorrect folding of the mutant peptide. However, the peptide is only 64 residues long and, once separated from the rest of the TrfA protein, may have little structure. In general β2-binding sites occur in regions of proteins with higher sequence and length variation than the rest of the protein, suggesting that they are not highly structured (7).
Hda, which was shown to be a suppressor of T1-associated cell lethality, also binds TrfA (17). However, as Kim et al. (17) pointed out, T1 interference with Hda's function alone is not sufficient to cause cell lethality, as Hda is not an essential protein in E. coli (5), nor is the negative regulation of DnaA by ATP hydrolysis through DnaA-Hda and β2 interaction essential to cell growth. Although the stimulation of ATP hydrolysis of DnaA is the predominant mechanism in E. coli for maintaining controlled initiation (6), it is not the only mechanism to regulate DnaA function in E. coli. Other biochemical processes such as the hemimethylated state of newly synthesized DNA (32), the SeqA protein to prevent DnaA from binding to oriC (28), and the titration of the DnaA protein at the datA locus (18, 31) are known to be operating in E. coli. cells to prevent extra initiation events.
A key unresolved issue is how the overexpression of Hda can suppress T1 toxicity, when overexpression of both T1 (16; this study) or Hda (1) alone is detrimental to host cells. In chromosome replication, β2 is critical to both initiation and elongation; successful negotiation of these phases relies on a balance between the levels of different β2-binding proteins and the stage of replication. Substantial overexpression of a protein or peptide that binds to β2 is detrimental to the cell (1, 2, 17), presumably by interfering with the initiation and/or elongation phases of DNA replication by competing with components of the replication system for β2 complexes. Currently, it is not clear which stage(s) of the process are the most susceptible to disruption by aberrant expression of molecules which bind to β2 at the major protein-protein-binding site. In vivo, expression of T1 appears to be very much more toxic than overexpression of Hda (1, 16), and toxicity of the T1 peptide is relieved with a small increase in the expression of Hda (17). DnaA-Hda-β2 forms a productive complex at the initiation of chromosome replication, which promotes the switching to elongation (24), while T1-β2 interaction is nonproductive. A small increase in the expression of Hda may compete with the T1 peptide and overcome a block in initiation, or at the transition from initiation of replication to elongation, without leading to toxicity itself. High-level overexpression of Hda may have additional impacts outside of the interaction of proteins with β2 (1).
β2 is harnessed by many DNA polymerases and enzymes which are active during initiation of DNA replication and replication elongation as well as the SOS (DNA damage) response (27, 42). The emerging view is that β2 is more of a ringmaster of the genome in that it acts as a mobile platform upon which a variety of different proteins may interact. Thus, β2's many roles include cell cycle control via its interaction with DnaA and Hda. The multiplicity of β2-binding proteins in one cell, all predicted to bind at the same site on β2, raise questions concerning the nature of the regulation of these interactions. β2 expression in the cell has been shown to be quite abundant (about 5,000 dimers per cell) (11). Factors influencing competition for the site on β2 may be (i) the site on the duplex DNA at which β2 is located, e.g., a lesion, a mismatch site, or a replicative primer; (ii) the dynamics of other proteins in proximity to the site, as part of transient active complexes or stalled, dissembling complexes; and (iii) the intrinsic “competitiveness” of the β2-binding motif of the protein.
We need to account for the observations that overexpression of both the longer TrfA peptide T1-2, containing amino acids 99 to 234 of TrfA, and the entire TrfA protein are not toxic to cells. Both of these proteins and the mutant peptide T1 (ΔLF) can accumulate in cells at high levels without affecting cell viability (data not shown). This may be viewed as differential affinity in the protein-protein interaction, which is related to accessibility of sequences to the surface of β2. We suggest that the binding of T1 to β2 is greater than the binding of T1-2 or TrfA to β2, perhaps due to the folding or lack thereof of the shorter peptide. However, this hypothesis remains to be tested. Alternatively, the binding site on β2 may be more accessible to the T1 peptide due to its small size.
It has been demonstrated that Hda interacts with TrfA (16), and it is also possible that T1-2 binds Hda, but what about T1 or T2? The major feature of T2 is that it contains part of the DNA-binding domain of TrfA, which is part of a highly conserved fold widely represented in plasmid Rep proteins (B. D. Dalrymple, unpublished results). Consequently, it is unlikely that the Hda interaction sequence is in this region of the protein unless it is across T1/T2 or in the first 20 amino acids or so of T2.
Acknowledgments
We thank Roger Pearson for advice on protein purification and Susan Briscoe for assistance with plate assays.
REFERENCES
- 1.Banack, T., N. Clauson, N. Ogbaa, J. Villar, D. Olver, and W. Firshein. 2005. Overexpression of the Hda DnaA-related protein in Escherichia coli inhibits multiplication, affects membrane permeability, and induces the SOS response. J. Bacteriol. 187:8507-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, R. E., E. B. Gottlin, D. J. Christensen, and P. T. Hamilton. 2003. Intracellular expression of peptide fusions for demonstration of protein essentiality in bacteria. Antimicrob. Agents Chemother. 47:2875-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnouf, D. Y., V. Olieric, J. Wagner, S. Fujii, J. Reinbolt, R. P. Fuchs, and P. Dumas. 2004. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competitive between replicative and translesion polymerases. J. Mol. Biol. 30:1187-1197. [DOI] [PubMed] [Google Scholar]
- 5.Camara, J. E., K. Skarstad, and E. Crooke. 2003. Controlled initiation of chromosome replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 185:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camara, J. E., A. M. Breier, T. Brendler, S. Austin, N. R. Cozzarelli, and E. Crooke. 2005. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 6:736-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohrmann, P. R., and C. S. McHenry. 2005. A bipartite polymerase-processivity factor interaction: only the internal β binding site of the α subunit is required for processive replication by the DNA polymerase III holoenzyme. J. Mol. Biol. 350:228-239. [DOI] [PubMed] [Google Scholar]
- 9.Fang, F. C., and D. R. Helinski. 1991. Broad-host-range properties of plasmid RK2: importance of overlapping genes encoding the plasmid replication initiation protein TrfA. J. Bacteriol. 173:5861-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeruzalmi, D., O. Yurieva, Y. Zhao, M. Young, J. Stewart, M. Hingorani, M. O'Donnell, and J. Kuriyan. 2001. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417-428. [PubMed] [Google Scholar]
- 11.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosome replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, T., and K. Sekimizu. 1999. Inactivation of Escherichia coli DnaA protein by DNA polymerase III and negative regulations for initiation of chromosomal replication. Biochimie 81:835-840. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, T., K. Fujimitsu, and T. Ogawa. 2001. Multiple pathways regulating DnaA function in Escherichia coli: distinct roles for DnaA titration by the datA locus and the regulatory inactivation of DnaA. Biochimie 83:13-17. [DOI] [PubMed] [Google Scholar]
- 14.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, P. D., T. M. Rosche, and W. Firshein. 2000. Identification of a potential membrane targeting region of the replication initiation protein (TrfA) of broad host range plasmid RK2. Plasmid 43:214-222. [DOI] [PubMed] [Google Scholar]
- 17.Kim, P. D., T. Banack, D. M. Lerman, J. C. Tracy, J. E. Camara, E. Crooke, D. Oliver, and W. Firshein. 2003. Identification of a novel membrane-associated gene product that suppresses toxicity of a TrfA peptide from plasmid RK2 and its relationship to the DnaA host initiation protein. J. Bacteriol. 185:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication inhibition by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittel, B. L., and D. R. Helinski. 1991. Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc. Natl. Acad. Sci. USA 88:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, X.-P., R. Onrust, M. O'Donnell, and J. Kuriyan. 1992. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425-437. [DOI] [PubMed] [Google Scholar]
- 21.Konieczny, I., and D. R. Helinski. 1997. Helicase delivery and activation by DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J. Biol. Chem. 272:33312-33318. [DOI] [PubMed] [Google Scholar]
- 22.Konieczny, I., K. S. Doran, D. R. Helinski, and A. Blasina. 1997. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol. Chem. 272:20173-20178. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg, A., and T. A. Baker. 1992. DNA replication. Freeman Press, New York, N.Y.
- 24.Kurz, M., B. Dalrymple, G. Wijffels, and K. Kongsuwan. 2004. Interaction of the sliding clamp β-subunit and Hda, a DnaA related protein. J. Bacteriol. 186:3508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenne-Samuel, N., J. Wagner, H. Etienne, and R. P. Fuchs. 2002. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 3:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase and DNA polymerase I. Proc. Natl. Acad. Sci. USA 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López de Saro, F. J., R. E. Georgescu, M. F. Goodman, and M. O'Donnell. 2003. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 22:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA, a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 29.Naktinis, V., J. Turner, and M. O'Donnell. 1996. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell 84:137-145. [DOI] [PubMed] [Google Scholar]
- 30.Oakley, A. J., P. Prosselkov, G. Wijffels, J. L. Beck, M. C. J. Wilce, and N. E. Dixon. 2003. Flexibility revealed by the 1.85 Å crystal structure of the β2 sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. D 59:1192-1199. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, T., Y. Yamada, T. Kuroda, T. Kishi, and S. Moriya. 2002. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44:1367-1375. [DOI] [PubMed] [Google Scholar]
- 32.Russell, D. W., and N. D. Zinder. 1987. Hemimethylation prevents DNA replication in E. coli. Cell 50:1071-1079. [DOI] [PubMed] [Google Scholar]
- 33.Skarstad, K., and E. Boye. 1994. The initiator protein DnaA: evolution, properties, and function. Biochim. Biophys. Acta 1217:111-120. [DOI] [PubMed] [Google Scholar]
- 34.Smith, C. A., and C. M. Thomas. 1984. Nucleotide sequence of the trfA gene of broad host range plasmid RK2. J. Mol. Biol. 175:251-262. [DOI] [PubMed] [Google Scholar]
- 35.Stukenberg, P. T., P. S. Studwell-Vaughan, and M. O'Donnell. 1991. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266:11328-11334. [PubMed] [Google Scholar]
- 36.Su'etsugu, M., M. Takata, T. Kubota, Y. Matsuda, and T. Katayama. 2004. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells 9:509-522. [DOI] [PubMed] [Google Scholar]
- 37.Sutton, M. D. 2004. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J. Bacteriol. 286:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton, M. D., I. Narumi, and G. C. Walker. 2002. Posttranslational modification of the umuD-encoded subunit of Escherichia coli DNA polymerase V regulates its interactions with the β processivity clamp. Proc. Natl. Acad. Sci. USA 99:5307-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, J., M. M. Hingorani, Z. Kelman, and M. O'Donnell. 1999. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 18:771-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. P. Fuchs. 2000. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wijffels, G., B. Dalrymple, P. Prosselkov, K. Kongsuwan, V. C. Epa, P. E. Lilley, S. Jergic, J. Buchardt, S. E. Brown, P. F. Alewood, P. A. Jennings, and N. E. Dixon. 2004. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry 43:5661-5671. [DOI] [PubMed] [Google Scholar]
- 42.Vivona, J. B., and Z. Kelman. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167-172. [DOI] [PubMed] [Google Scholar]