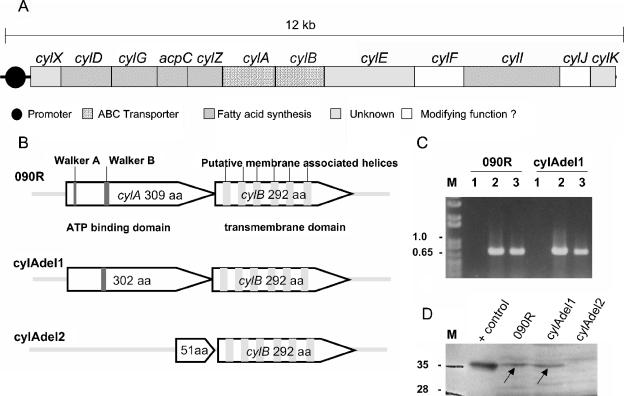
FIG. 1.
(A) Genomic map of the cyl gene cluster. Depicted are the 12 genes belonging to the cluster. Homologies of the deduced proteins to genes with known function in the GenBank database and the putative promoter region are indicated. Genes that are indicated as open boxes display homologies to genes encoding enzymes such as an aminomethyltransferase (cylF annotated in genome sequencing project AAJS01000020 as belonging to protein family PF01571) and a glycosyl transferase (cylJ harbors the conserved motif COG1819 of glycosyl transferases), which may modify the structure of an assembled molecule. (B) Graphic representation of the deletion mutants of cylA. Indicated are the Walker A and Walker B sites of cylA and the six putative transmembrane helices of cylB in the wild-type strain. The truncated versions of CylA in both mutants are depicted. (C) Transcription analysis of cylE by RT-PCR. Strains 090R and cylAdel1 were grown to mid-logarithmic phase, and 50 ng of total RNA was used as a template for reverse transcription. The RT primer annealed at nucleotides 765 to 748 of cylE, and the subsequent PCR amplified nucleotides 39 to 765 of cylE (lane2). Chromosomal DNA served as a positive control (lane 3), and 50 ng of RNA that had not been subjected to an RT reaction (RT inactivated) was used to control for DNA contamination of RNA samples (lane 1). M, molecular mass marker (kb). (D) Western blot analysis of bacterial membrane fractions with anti-CylA antibodies. S. agalactiae wild-type strain 090R and the mutant strains cylAdel1 and cylAdel2 were grown to late logarithmic phase in THB supplemented with 1% proteose peptone. Subcellular fractions of all strains were prepared as detailed in Materials and Methods. Western blot analysis was carried out with polyclonal anti-cylA (rabbit) antibodies at a dilution of 1:1,000. M, molecular mass marker; lane 1, 2 μg of recombinant CylA (+control). For lanes 2 to 4, 40 μg of membrane fractions was applied to each lane, as follows: lane 2, 090R, lane 3, cylAdel1; lane 4, cylAdel2.