Figure 1. Hsp104 catalyzes or abolishes the de novo assembly of Sup35 and Ure2 fibers.
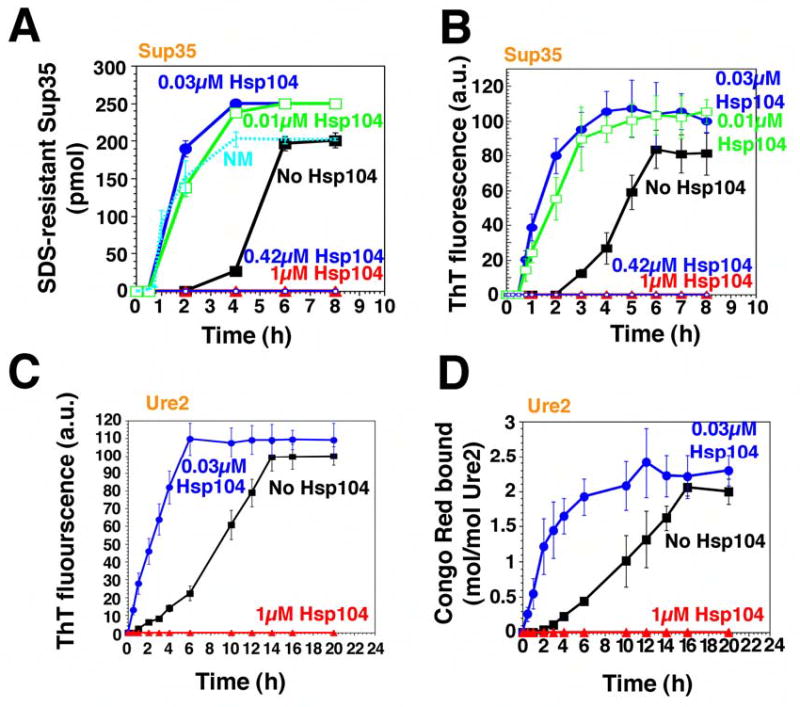
(A, B) Kinetics of unseeded, rotated (80rpm) Sup35 (2.5μM) polymerization without or with Hsp104 (0.01–1μM) plus ATP (5mM). Fiber assembly was assessed by the formation of SDS-insoluble Sup35 (A) and ThT fluorescence (B). In (A), the kinetics of unseeded, rotated (80rpm) NM (2.5μM) fibrillization is also shown. Values represent means±SD (n=5).
(C, D) Kinetics of unseeded, rotated (80rpm) Ure2 (2.5μM) polymerization without or with Hsp104 (0.03μM or 1μM), plus ATP (5mM). Fibrillization was measured by ThT fluorescence (C) or CR binding (D). Values represent means±SD (n=3).
