Figure 6. Hsp104 couples ATP hydrolysis to Ure2 prion fragmentation.
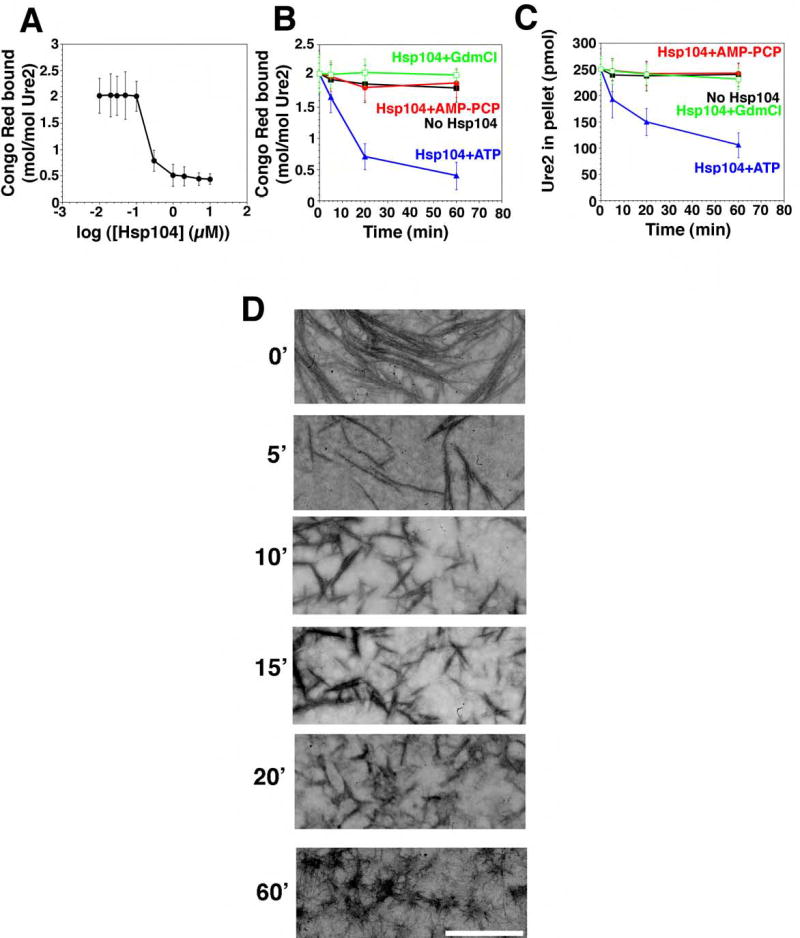
(A) Ure2 (2.5μM) was incubated for 16h with rotation (80rpm) to generate fibers and then incubated with Hsp104 (0.01–10μM) plus ATP (5mM) for 60min at 25°C. Disassembly was assessed by CR binding. Values represent means±SD (n=3). (B, C) Kinetics of Hsp104 (2μM) induced Ure2 fiber (2.5μM monomer) disassembly in the presence of either ATP (5mM), ATP (5mM) plus GdmCl (20mM), or the non-hydrolyzable analog AMP-PCP (5mM) (an alternative to AMP-PNP). Disassembly was monitored by CR binding (B) or sedimentation analysis (C). Values represent means±SD (n=3).
(D) Ure2 fibers (2.5μM monomer) were incubated with Hsp104 (2μM) plus ATP (5mM) for 0–60min, and at various times reactions were processed for EM. Scale bar, 0.25μm.
