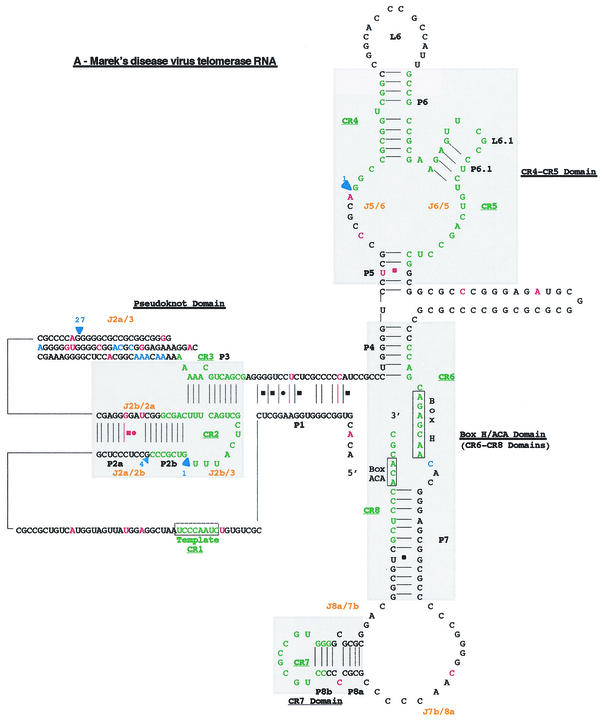
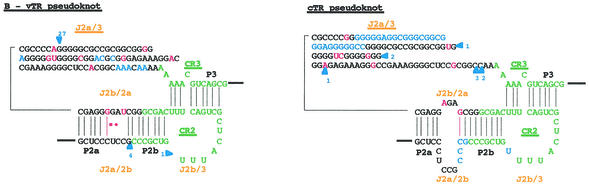
FIG. 4.
Proposed secondary structure of the MDV. Comparison of the overall proposed secondary structure of MDV and cTR (A) and their pseudoknot domains structures (B) are based on the vertebrate TR structure published by Chen et al. (9). Paired regions (P) are numbered from 5′ to 3′ as P1 to P8. Junction regions (J) between two paired regions are named with reference to the flanking paired regions and are given in orange type. The four universal structural domains conserved in all vertebrate TRs are shaded in gray and labeled. The template region, box H, and box ACA motifs are labeled, and their conserved nucleotides are boxed. Universal base pairing (according to the Watson-Crick scheme) is represented by dashes, whereas G-U pairs and noncanonical A-C pairs are indicated by squares and circles, respectively. Mutations in the vTR sequence are in red, as is additional base pairing in the vTR structure. Additional nucleotides in the vTR sequence are shown in blue, and deletion regions are indicated by blue arrows showing the location and the nucleotide length of the deletion.