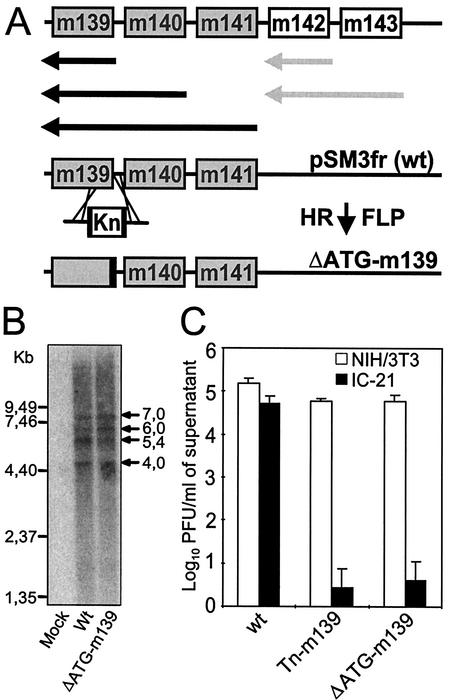
FIG. 4.
Construction, analysis, and growth of mutant ΔATG-m139. (A) Transcript map of genes m139 to m143 and construction of the ATG deletion mutant of m139. Black and grey arrows denote the major transcripts of genes m139 to m141 and genes m142 and m143, respectively, according to Hanson et al. (18). Deletion of the first two ATGs of m139 was achieved by site-directed mutagenesis with a linear PCR fragment containing a kanamycin resistance gene (Kn) flanked by FRT sites (black boxes) and by 50 nucleotides homologous to the up- and downstream sequences. After insertion of the kanamycin resistance gene by homologous recombination (HR), the kanamycin cassette was excised by FLP recombinase, generating the ΔATG-m139 mutant genome. (B) Northern blot analysis of the m139 to m141 transcripts. NIH 3T3 cells were infected at 5 PFU/cell with wild-type (wt) or ΔATG-m139 MCMV, and mRNA was isolated as described in Materials and Methods. The probe used for detection of these transcripts binds to the m140 ORF. (C) Growth of the corresponding ΔATG-m139 MCMV mutant in NIH 3T3 fibroblasts and IC-21 macrophages. Monolayers of NIH 3T3 fibroblasts and IC-21 macrophages were infected at an MOI of 0.1. The titers of cell-free viruses were determined on day 6 postinfection.