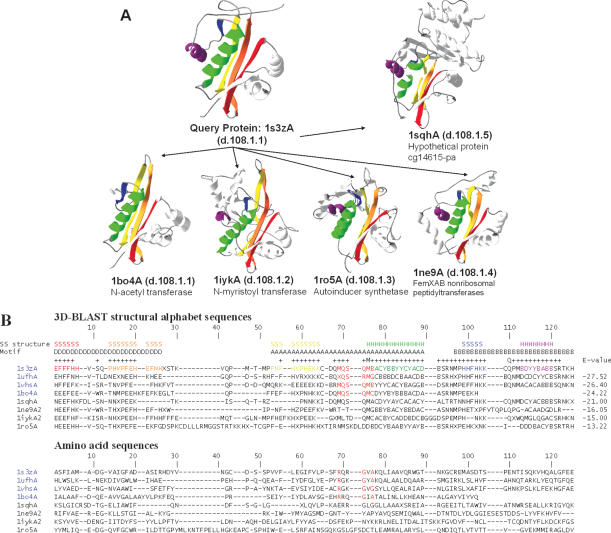
Figure 5.
Multiple structure alignment and multiple sequence alignment of 3D-BLAST search results using the aminoglycoside 6′-NAT from S.enterica as the query. (A) The structural recurrences of five homologous proteins from the NAT superfamily are colored. Each structure is shown as a continuous color spectrum from red through orange, yellow, green and blue to violet. The PDB code and SCOP sccs are also indicated. (B) Sequence alignments of both structural alphabet and amino acid sequences of eight proteins. The location of secondary structures [i.e. strand (S) and helix (H)] and the motifs D, A and B are indicated. Motif A presents an invariant Arg-X-X-Gly-X-Gly/Ala segment (red columns) for four proteins (PDB code 1s3zA, 1ufhA, 1vhsA and 1bo4A) of the NAT family, and the secondary structures are obtained from DSSP based on the query protein. The structural alphabet sequences are more conserved than the amino acid sequences. The E-value between the query and homologous proteins from the 3D-BLAST results is given for each sequence.