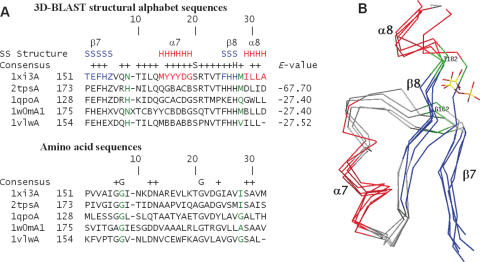
Figure 6.
(A) Multiple sequence alignments and (B) multiple structure alignments resulting from a 3D-BLAST search using thiamine phosphate pyrophosphorylase from P.furiosus as the query. The aligned proteins are 1 × i3A and 2tpsA (thiamine phosphate synthase superfamily), 1qpoA (quinolinic acid phosphoribosyltransferase superfamily), 1w0mA (triosephosphate isomerase superfamily) and 1vlwA (aldolase superfamily). The alignment and superimposition region, a common phosphate-binding site of these five proteins, is from β7 to α8. The secondary structures of these proteins are indicated in red (helices) and blue (strands), whereas the remaining structures are shown in gray. The residues involved in phosphate-binding are indicated in green.