Abstract
The DNA methyltransferases, Dnmts, are the enzymes responsible for methylating DNA in mammals, which leads to gene silencing. Repression by DNA methylation is mediated partly by recruitment of the methyl-CpG-binding protein MeCP2. Recently, MeCP2 was shown to associate and facilitate histone methylation at Lys9 of H3, which is a key epigenetic modification involved in gene silencing. Here, we show that endogenous Dnmt3a associates primarily with histone H3-K9 methyltransferase activity as well as, to a lesser extent, with H3-K4 enzymatic activity. The association with enzymatic activity is mediated by the conserved PHD-like motif of Dnmt3a. The H3-K9 histone methyltransferase that binds Dnmt3a is likely the H3-K9 specific SUV39H1 enzyme since we find that it interacts both in vitro and in vivo with Dnmt3a, using its PHD-like motif. We find that SUV39H1 also binds to Dnmt1 and, consistent with these interactions, SUV39H1 can purify DNA methyltransferase activity from nuclear extracts. In addition, we show that HP1β, a SUV39H1-interacting partner, binds directly to Dnmt1 and Dnmt3a and that native HP1β associates with DNA methyltransferase activity. Our data show a direct connection between the enzymes responsible for DNA methylation and histone methylation. These results further substantiate the notion of a self-reinforcing repressive chromatin state through the interplay between these two global epigenetic modifications.
INTRODUCTION
The enzymatic methylation of cytosine within the context of a simple dinucleotide site, CpG, is an epigenetic process that is essential for mammalian development. DNA methylation plays a key role in controlling several biological processes, such as X chromosome inactivation, genomic imprinting, genomic stability and chromatin structure, most likely as a result of its well-documented repressive effect on gene transcription (1).
Methylation of CpG dinucleotides in mammals is carried out by three active DNA methyltransferases, Dnmt1, Dnmt3a and Dnmt3b (2). Disruption of Dnmt1 in mice results in early embryonic lethality and severe genomic hypomethylation (3). Mice disrupted for Dnmt3a or Dnmt3b undergo abnormal mammalian development and have a loss of genomic DNA methylation in pericentromeric repeats (4). The mechanisms by which DNA methylation brings about transcriptional repression have been the subject of intense research over the last few years. It has been demonstrated that gene silencing can occur through the action of proteins that bind methylated CpG sequences (methyl-CpG-binding domain proteins, MBDs), which are components or are recruited to methylated DNA by histone deacteylases (HDAC), which remove acetyl groups from the tail of histones and help maintain nucleosomes in a compacted and transcriptionally silent state (5). A more direct connection between DNA methylation and deacetylation has more recently been elucidated, where the Dnmt enzymes directly recruit HDAC activity to silence gene expression (6–10). Thus, DNA methylation and histone deacetylation function through a common mechanistic pathway to repress transcription.
More recently, DNA methylation has been linked to another chromatin-associated transcriptional repression process involving the methylation of Lys9 of histone H3. It was found that mutations of the gene dim-5 in Neurospora crassa or kryptonite in Arabidopsis thaliana results in loss of DNA methylation in these organisms. Interestingly, dim-5 and kryptonite were shown to encode histone H3 Lys9 (H3-K9) methyltransferases (11,12). Thus these data revealed that, in Neurospora and Arabidopsis, histone methylation can direct DNA methylation. The mechanisms underlying this dependence of DNA methylation on histone modification are still unclear. In Arabidopsis the adaptor protein LHP1, which binds with high affinity to histone H3 when methylated at Lys9, was found to interact with the CMT3 DNA methyltransferase. Hence, it was proposed that LHP1 would recruit CMT3 to DNA that has to be methylated and thereby histone methylation would influence DNA methylation (12).
Not only can histone modification induce DNA methylation signals but the reverse can also occur. New findings in mammals reveal that the MBD repressor MeCP2 associates with histone H3 Lys9 methyltransferase activity and delivers this chromatin modification to a DNA methylated gene that it regulates (13). Thus, DNA methylation can also feed back on lysine methylation, suggesting that these two global epigenetic modifications could act together to perpetuate and maintain a repressed chromatin state.
In the present study, we show that the DNA methyltransferases themselves are directly linked to histone methylation. We find that endogenous Dnmt3a purifies histone methyltransferase activities that primarily modify Lys9 of H3 but also to a lesser extent Lys4 of H3. The SUV39H1 H3-K9 methylase is likely the enzyme responsible for the Dnmt3a-associated H3-K9 methyltransferase activity since we find that it binds both in vitro and in vivo to Dnmt3a, through its PHD-like motif. The DNA methyltransferase Dnmt1 also binds to SUV39H1. Both Dnmt1 and Dnmt3a can contact HP1β directly. Finally, we find that SUV39H1 as well as HP1β associate with DNA methyltransferase activity. These data establish a much more direct connection between DNA methylation and histone modification and further support the concept of interdependent processes between these two epigenetic layers.
MATERIALS AND METHODS
Plasmids
The various pGEX vectors for Dnmt3a as well the Myc-tagged full-length mammalian expression construct of Dnmt3a have been described previously (7). The following plasmids have been described before: pCDNA3.1/Myc-His Dnmt1 (7), pcDNA3.1-GAL4 Dnmt3a 490–582 (8), pGEX-HP1β (14), pcDNA3.1-HA SUV39H1 (15), pcDNA3.1-GAL4 SUV39H1 (15) and pGEX-Rb (15). The various glutathione S-transferase (GST)–Dnmt1 fusion plasmids were cloned from corresponding pET28 constructs (16) and were a kind gift from Dr Albert Jeltsch. The pET16b His-tagged HP1β constructs were a kind gift from Ernest Laue. The pMAL and pGEX expression vectors for SUV39H1 were subcloned by PCR using appropriate sets of primers. We verified all constructs by DNA sequencing.
Cell culture and transfections
Monolayer cultures of U2OS and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum (Clontech) and grown at 37°C, 5% CO2. Transfections were carried out using the calcium phosphate method.
GST and maltose-binding protein (MBP) fusion proteins, in vitro translations and pull-down assays
We expressed GST and GST fusion proteins in Escherichia coli XA90 using the pGEX (Pharmacia) vector system and purified proteins from crude bacterial lysates according to the manufacturer’s instructions. MBP and MBP fusion proteins were expressed in E.coli XA90 using the pMAL (NEB) vector system and purified according to the manufacturer’s instructions. In vitro transcription–translation was performed using the TNT system (Promega). Dnmt1 and Dnmt3a were translated in vitro from pCDNA3.1/Myc-His Dnmt1 or pCDNA3.1/Myc-His Dnmt3a, respectively. We performed pull-down assays essentially as described previously (17).
Direct interaction assays using His-tagged proteins
We expressed histidine-tagged full-length HP1β or the HP1β chromo-shadow domain as described before (8). Purified His-HP1β or HP1β chromo-shadow domain were incubated with GST fusion proteins for 2 h at 4°C in IPH buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% v/v Nonidet-P40, 5 mM EDTA). Reactions were washed four times in IPH buffer and resolved by SDS–PAGE. Gels were blotted onto nitrocellulose membranes and subjected to western blot analysis using anti-His antibody (BMG-His-1; Roche).
GST pull-downs and immunoprecipitation for histone methyltransferase assays
Pull-down assays were done by incubating GST fusion proteins prebound to glutathione beads with HeLa nuclear extracts (50 µl) in RIPA buffer (200 µl, 50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P40 and 0.1% SDS). The incubation was performed at 4°C for 12 h. The beads were then washed four times in 1 ml of RIPA buffer before processing to histone methyltransferase assays. For immunoprecipitations prior to histone methyltransferase assays, antibodies against Dnmt3a (a gift from En Li) were used. HeLa nuclear extracts (50 µl) (Computer Cell Culture Centre, Belgium) were incubated with the relevant antibodies overnight at 4°C. Antibody complexes were collected on protein A/G–Sepharose beads, washed four times in RIPA buffer and assayed for histone methyltransferase assays.
Histone methyltransferase assays and protein sequencing
Precipitations from pull-downs or immunoprecipitations from HeLa nuclear extract were incubated with 10 µg of histones (Sigma) and 2 µl S-adenosyl-l-[methyl-3H]methionine (67 Ci mmol–1) (Amersham Pharmacia) in buffer MAB (50 mM Tris pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM 2-mercaptoethanol and 250 mM w/v sucrose) at 30°C for 4 h. The reaction products were then resolved by SDS–PAGE, western blotted and autoradiographed. The position of radiolabelled histone H3 was identified by its size using radiolabelled molecular weight markers (Amersham) as well as by comparing the autoradiography with the gel stained with Ponceau. For N‐terminal sequencing, radiolabelled histone H3 was blotted to PVDF (Millipore) and sequenced by Edman degradation (Protein Sequencing Facility, University of Cambridge). Amino acid fractions were analysed for the presence of tritium by scintillation counting.
DNA methyltransferase assays
DNA methyltransferase assays were performed as described before (7). The HP1β antibody was a gift of R. Laskey.
Immunoprecipitations and western blot analysis
U2OS cells were transiently transfected in culture dishes (10 cm diameter) with 6 µg of each expression vector. Cells were harvested 24 h post-transfection, lysed in 400 µl of IPH lysis buffer (18) at 4°C for 30 min and debris removed by centrifugation. Immunoprecipitations and western blotting were then carried out as described previously (8). Anti-GAL4 (5C1; Santa Cruz), anti-HA (12C5A; Roche) or anti-Myc (9E10; Roche) antibodies were used.
RESULTS
Dnmt3a associates with histone methylase activity that is specific for H3
Our recent finding that the methyl-CpG-binding protein MeCP2 associates with histone methyltransferase activity (13) prompted us to ask whether the DNA methyltransferases themselves could be linked to histone methylation. To test this possibility, we first investigated if antibodies against the DNA methyltransferase Dnmt3a could precipitate histone methyltransferase activity from untransfected cells. We measured associated histone methyltransferase activity in Dnmt3a immunoprecipitates from HeLa cells using 3H-labelled S-adenosylmethionine as a potential methyl group donor and incubated with a mixture of bulk histones as a substrate. Histones showing transfer of the labelled methyl groups were then separated by SDS–PAGE and visualised by western blotting and autoradiography. As shown in Figure 1A, two distinct antibodies raised against Dnmt3a purified an activity that methylates histone H3 from nuclear extracts (lanes 1 and 3), whereas their respective preimmune sera failed to do so (P.I., lanes 2 and 4). Of note, the other histones, H2A, H2B and H4, were not methylated after precipitation with the Dnmt3a antibodies. Thus, endogenous Dnmt3a co-immunoprecipitates histone methyltransferase activity that is specific for histone H3.
Figure 1.
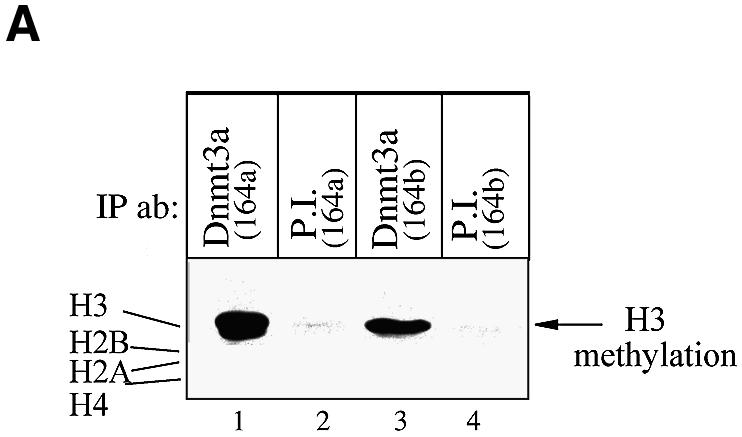
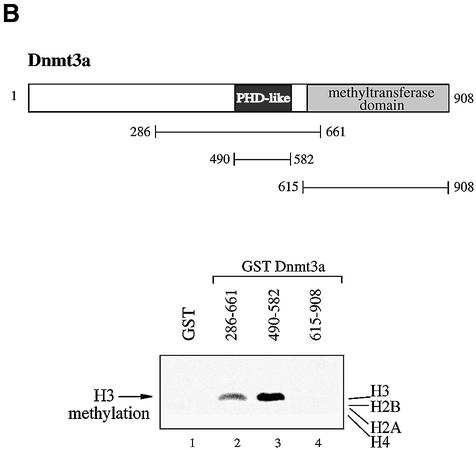
Endogenous Dnmt3a purifies histone methyltransferase activity specific for H3. (A) Endogenous Dnmt3a associates with H3 methyltransferase activity. HeLa nuclear extracts were immunoprecipitated with either antibodies against Dnmt3a (164a or 164b; lanes 1 and 3) or their respective preimmune sera (lanes 2 and 4). After washing, the immune complexes where tested for histone methyltransferase activity using bulk histones as substrate. The reaction products were then analysed by SDS–PAGE followed by western blotting and autoradiography. The radiolabelled H3 is indicated by an arrow on the right. (B) Dnmt3a binds H3 methylase activity through its conserved PDH-like motif. On top is depicted a schematic representation of Dnmt3a with the different domains that were fused to GST. Numbers indicate amino acid residues. HeLa nuclear extracts and equivalent amounts of the indicated GST fusions bound to Sepharose beads were incubated with bulk histones as substrate. The beads were then washed and assayed for histone methyltransferase activity.
To define the domains within Dnmt3a that are required for the association with HMTase activity, we used bacterially expressed GST fusion proteins containing various domains of Dnmt3a. The GST fusions on beads were incubated with HeLa nuclear extracts and assayed for histone methyltransferase activity. We found that amino acids encompassing the conserved PHD-like motif of Dnmt3a (residues 286–661) or bearing only this motif (490–582) associate efficiently with HMTase activity (Fig. 1B, lanes 2 and 3). This association of enzymatic activity was specific since the control GST alone or another region of Dnmt3a, the catalytic C-terminal domain (residues 615–908), failed to bind HMTase activity (Fig. 1B, lanes 1 and 4).
Dnmt3a is bound to HMTases that methylate primarily H3-K9 and to a lesser extent H3-K4
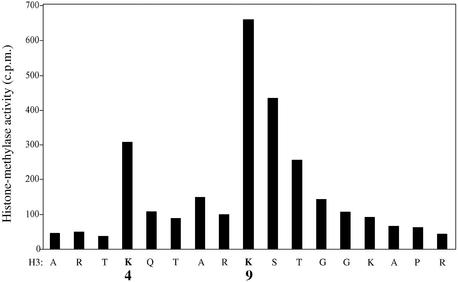
We next wished to determine the residues of histone H3 that are modified by the Dnmt3a-interacting HMTase(s). To this end, histone H3 radioactively methylated by the Dnmt3a-associated histone methylase(s) was used for Edman degradation. Protein sequencing was then performed on each fraction and radioactivity counted using a liquid scintillation counter. As shown in Figure 2, the Dnmt3a-associated HMTase activities preferentially methylate Lys9 of histone 3 (H3-K9) and to a lesser extent histone H3 at Lys4 (H3-K4). Methylation of lysine residues on histone H3 is known to occur on K4 and K9, and distinct enzymes have been identified that methylate these sites (19). It is therefore likely that two different sets of histone methyltransferases are associated with Dnmt3a, one set that weakly methylates Lys4 and another one that more efficiently adds methyl groups to Lys9 of histone H3. While the nature of the Dnmt3a-interacting H3-K4 HMTase is currently unknown, we present data (see below) regarding the identity of the histone Lys9 methyltransferase that associates with the DNA methyltransferases.
Figure 2.
The Dnmt3a-associated HMTases methylate primarily H3-K9 and to a lesser extent H3-K4. Histone H3 labelled by Dnmt3a-associated HMTase activities were subjected to Edman degradation and each fraction was then counted by liquid scintillation counting.
The H3-K9 methyltransferase SUV39H1 associates with Dnmt3a and Dnmt1
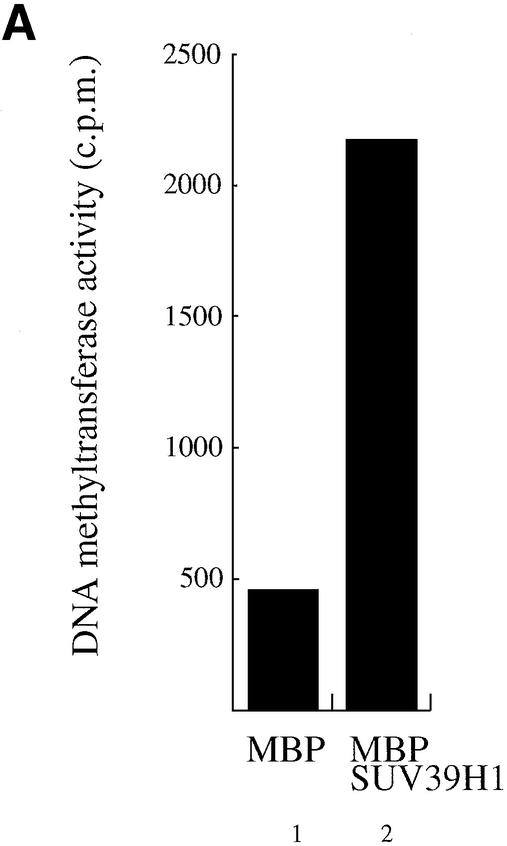
Dnmt3a binds a HMTase that efficiently modifies Lys9 of histone H3 (see Figs 1 and 2). So far, five mammalian enzymes have been described to methylate H3-K9 (19). Among these, the first and best characterised H3-K9 HMTase is the SUV39H1 protein. SUV39H1 localises to transcriptionally silent heterochromatic sites (20) and mice lacking the Suv39h1/h2 methylases display impaired mammalian heterochromatin and genome stability (21). The role of SUV39H1 is not restricted to constitutive heterochromatin as it is also recruited to euchromatic genes, such as cyclin E, to silence gene expression (15). As a first hint to determine whether SUV39H1 is the H3-K9 HMTase that associates with Dnmt3a, we tested whether SUV39H1 could associate with DNA methyltransferase activity. To this end, SUV39H1 was fused to MBP and incubated with HeLa nuclear extracts, washed and assayed for DNA methyltransferase activity using an oligonucleotide substrate and 3H-labelled S-adenosyl-l-methionine as a methyl group donor. Incorporation of radioactivity was determined by liquid scintillation counting. As shown in Figure 3A, MBP–SUV39H1 precipitates significant DNA methyltransferase activity from HeLa nuclear extracts (lane 2), whereas MBP alone gave only background activity (lane 1).
Figure 3.
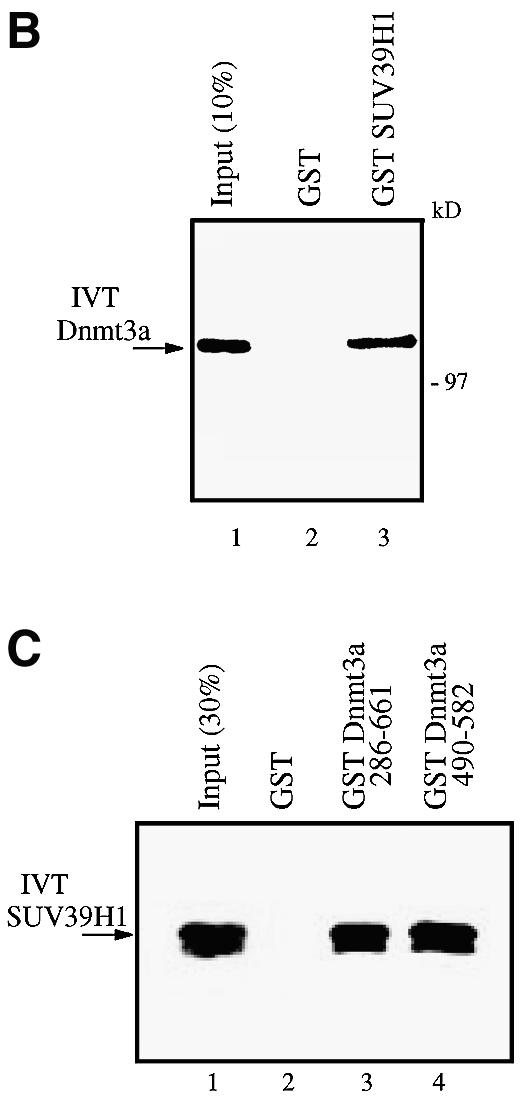
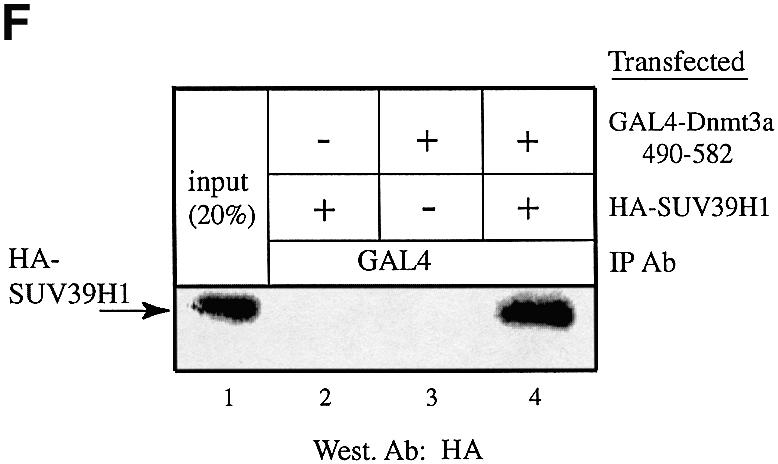
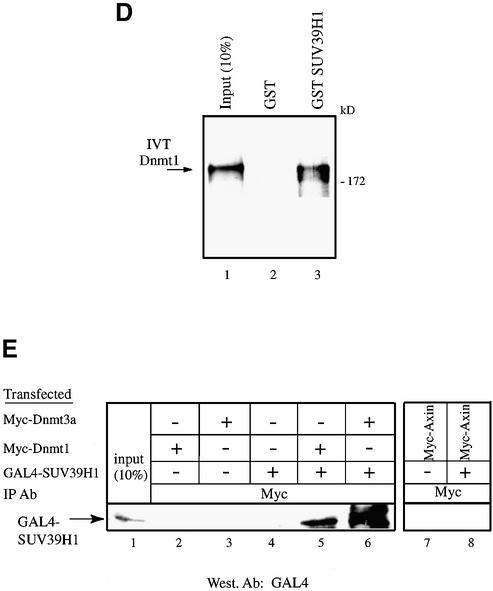
The SUV39H1 H3-K9 methylase binds to Dnmt3a and Dnmt1. (A) SUV39H1 associates with DNA methyltransferase activity from nuclear extracts. Equivalent amounts of MBP (lane 1) or MBP–SUV39H1 fusion proteins (lane 2) were incubated with HeLa nuclear extract, washed and assayed for DNA methyltransferase activity. Activity is given as c.p.m. of S-adenosyl-l-[methyl-3H]methionine incorporated into a hemi-methylated oligonucleotide substrate. Incorporation of radioactivity was determined by liquid scintillation counting. (B) SUV39H1 binds in vitro to Dnmt3a. GST or GST–SUV39H1 were incubated with in vitro translated [35S]Dnmt3a and subjected to GST pull-downs. Lane 1 shows 10% of the Dnmt3a input. (C) The PHD-like motif of Dnmt3a mediates in vitro the association with SUV39H1. GST or GST–Dnmt3a (encompassing or containing its PHD-like motif) were incubated with in vitro translated [35S]SUV39H1 and subjected to GST pull-downs. Lane 1 shows 30% of the SUV39H1 input. (D) SUV39H1 interacts in vitro with Dnmt1. GST pull-downs were done using GST or GST–SUV39H1 incubated with in vitro translated [35S]Dnmt1. Lane 1, [35S]Dnmt1 input (10%). (E) SUV39H1 co- immunoprecipitates with Dnmt3a and Dnmt1. U20S cells were transfected with the indicated plasmids. Whole cell extracts were precipitated with Myc antibody and the presence of GAL4–SUV39H1 in the immunoprecipitate was detected by western blot using anti-GAL4 antibody. The unrelated Myc–Axin was used as a negative control and was tested in a separate experiment than the other constructs (lanes 7 and 8). The input lane contains 10% of total extract. (F) Dnmt3a co-immunoprecipitates with HA–SUVAR39H1 using its PHD-like finger. GAL4–Dnmt3a was precipitated using a GAL4 antibody and the presence of HA–SUV39H1 was detected by western blot using anti-HA antibody. Lane 1, 20% of input.
Having shown that SUV39H1 associates with Dnmt enzymatic activity, we next wished to examine whether SUV39H1 can interact with the Dnmt3a DNA methyltransferase. Toward this end, we performed in vitro GST pull-downs using SUV39H1 fused to GST and incubated with in vitro translated (IVT) 35S-labelled Dnmt3a. As shown in Figure 3B (lane 3), IVT-Dnmt3a was efficiently pulled down by GST–SUV39H1. In contrast, no radiolabelled protein was pulled down by the GST protein alone (lane 2). Consistent with the finding that the PHD-like motif of Dnmt3a is required to associate with histone methyltransferase activity (Fig. 1B), GST pull-downs indicate that the PHD-like motif of Dnmt3a mediates the interaction with in vitro translated IVT-SUV39H1 (Fig. 3C).
The Dnmt enzymatic activity bound to SUV39H1 (see Fig. 3A) could be provided by Dnmt3a but also by other Dnmts, such as Dnmt1. It was therefore possible that SUV39H1 would not only interact with Dnmt3a but also with Dnmt1. We tested this possibility by performing pull-downs with GST–SUV39H1 and IVT-Dnmt1. As shown in Figure 3D, IVT-Dnmt1 could also be pulled down specifically by GST–SUV39H1 (Fig. 3D, lane 3). Thus, SUV39H1 interacts in vitro specifically with both Dnmt1 and Dnmt3a.
We next asked whether these interactions could also be demonstrated in vivo using a co-immunoprecipitation approach. Mammalian 293 cells were co-transfected with expression vectors for SUV39H1, which was GAL4 tagged (GAL4–SUV39H1), and Dnmt1 or Dnmt3a tagged with Myc (Myc–Dnmt1 and Myc–Dnmt3a, respectively). Following immunoprecipitation with anti-Myc antibody, the immune complexes were analysed by immunoblotting with anti-GAL4 sera. Figure 3E shows that Dnmt1 interacted specifically with SUV39H1 (lane 6) since no precipitate was detected in the absence of exogenous SUV39H1 (lane 3) or in the absence of transfected Dnmt1 (lane 4). Similarly, when GAL4– SUV39H1 was co-transfected together with Myc–Dnm3a, we detected an interaction between SUV39H1 and Dnmt3a (Fig. 3E, lane 5). In contrast, no association was observed when either construct was transfected alone (lanes 2 and 4). In addition, no precipitate was detected after transfection of SUV39H1 and an expression vector for an unrelated protein (Myc-tagged Axin, lanes 7 and 8), thus further demonstrating the specificity of the association between SUV39H1 and Dnmt1 or Dnmt3a. Similar co-immunoprecipitation experiments were performed using GAL4-tagged Dnmt3a 490–582, which contains its PHD-like motif, and HA-tagged SUV39H1. As shown in Figure 3F (lane 4), Dnmt3a 490–582 co-immunoprecipitated with SUV39H1. This is consistent with the finding presented in Figure 3B where GST Dnmt3a 490–582 mediates the interaction with in vitro translated SUV39H1. Taken together, our data indicate that SUV39H1 associates with Dnmt enzymatic activity and interacts both in vitro and in vivo with the Dnmt1 and Dnmt3a DNA methyltransferases.
The heterochromatin protein HP1 contacts Dnmt1 and Dnmt3a directly and associates in vitro and in vivo with DNA methyltransferase activity
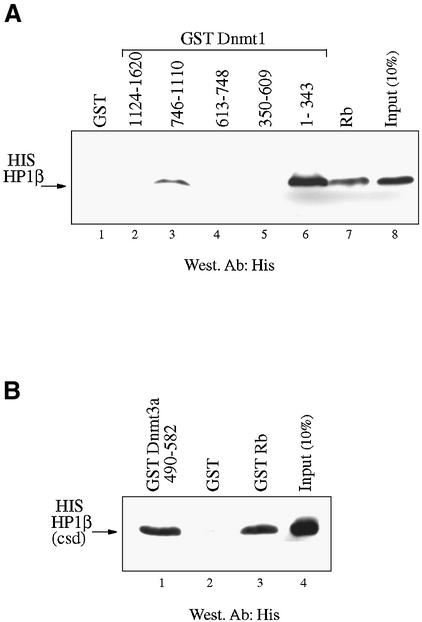
Recent data have demonstrated that SUV39H1 interacts with HP1 (22), a heterochromatic adaptor protein involved in epigenetic gene regulation and supranucleosomal chromatin structure. Mechanistically, it has recently been shown that methylation at Lys9 of histone H3 by the SUV39H1 enzyme creates a binding site for HP1 that recognizes the H3-K9 methyl epitope through its chromo domain (14,23). This mechanistic connection between SUV39H1 and HP1 prompted us to investigate whether HP1 could associate with Dnmt1 and/or Dnmt3a as well as with Dnmt enzymatic activities. We first examined the ability of HP1β, a member of the HP1 family, to bind Dnmt1 and/or Dnmt3a in direct GST pull-down assays by producing both HP1β and various Dnmt1 or Dnmt3a constructs as recombinant proteins in E.coli. As shown in Figure 4A, two N‐terminal regions of GST–Dnmt1 (residues 746–1110 and 1–343) bound histidine-tagged HP1β (lanes 3 and 6, respectively), whereas GST alone (lane 1) or other regions of Dnmt1 did not (lanes 2, 4 and 5). We used GST fused to the retinoblastoma protein (Rb), a protein known to bind to HP1β (15), as a positive control (Fig. 4A, lane 7). With respect to Dnmt3a, similar GST pull-downs also revealed that His-tagged HP1β [containing its chromo-shadow domain (csd), residues 104–185] bound directly to the PHD-like motif of Dnmt3a (Fig. 4B). Thus, both Dnmt1 as well as Dnmt3a can contact HP1β and these interactions are direct.
Figure 4.
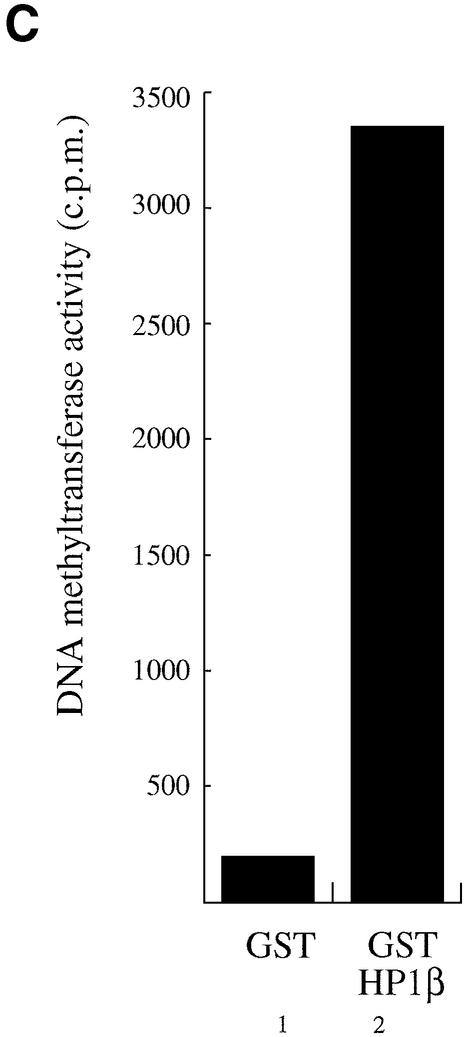
HP1β binds directly to Dnmt1 and Dnmt3a and interacts in vitro and in vivo with DNA methyltransferase activity. (A) HP1β directly contacts Dnmt1. GST, GST–Rb or the indicated GST–Dnmt1 fusions were incubated with histidine-tagged full-length HP1β. Direct binding was visualised by western blotting for HP1β using anti-His antibody (Roche). Input is 10% of total His-tagged HP1β in each assay. (B) HP1β binds directly to Dnmt3a. GST, GST–Rb or GST–Dnmt3a containing its PHD-like motif were incubated with histidine-tagged HP1β chromo-shadow domain (His-HP1β csd, residues 104–185). Western blotting was then performed using anti-His antibody. Input is 10% of total His-tagged HP1β chromo-shadow domain in each assay. (C) HP1β binds in vitro with DNA methyltransferase activity. Equivalent amounts of GST (lane 1) or GST–HP1β fusion proteins (lane 2) were incubated with HeLa nuclear extract. After extensive washing, the beads were assayed for DNA methyltransferase activity. Activity is given as c.p.m. of S-adenosyl-l-[methyl-3H]methionine incorporated into a hemi-methylated oligonucleotide substrate. Incorporation of radioactivity was determined by liquid scintillation counting. (D) Endogenous HP1β purifies DNA methyltransferase activity from nuclear extracts. HeLa nuclear extracts were immunoprecipitated with preimmune serum (lane 1) or an anti-HP1β antibody (lane 2). Immunoprecipitates were then assayed for DNA methyltransferase activity.
We then evaluated the ability of HP1β fused to GST to purify DNA methyltransferase activity from HeLa nuclear extracts. Enzymatic activity was measured as radioactivity of S-adenosyl-l-[methyl-3H]methionine incorporated into an oligonucleotide substrate. Figure 4C (lane 2) indicates that GST–HP1β associates with DNA methyltransferase activity from nuclear extracts. This association of enzymatic activity was specific since the control GST alone gave background only (Fig. 4C, lane 1).
Next, we asked whether endogenous HP1β could purify DNA methyltransferase activity from nuclear extracts. To test this, we immunoprecipitated native HP1β with a HP1β-specific antibody followed by Dnmt enzymatic assay. As shown in Figure 4D, a significant amount of DNA methyltransferase activity was purified using an anti-HP1β antibody (lane 2), whereas control immunoprecipitation with preimmune serum (P.I., lane 1) showed background activity. These results demonstrate that native HP1β associates with DNA methyltransferase activity.
DISCUSSION
Here we report an association between the DNA methyltransferases Dnmt1 and Dnmt3a and the SUV39H1–HP1 histone methylation system. We show that Dnmt3a associates primarily with histone H3 Lys9 methyltransferase activity but also to a lesser extent with H3 Lys4 activity. The H3 Lys9 enzymatic activity is likely provided by the H3-K9 HMTase SUV39H1 since we observed that it binds both in vitro and in vivo to Dnmt3a. We find that SUV39H1 also binds to Dnmt1 and consistent with these associations, SUV39H1 purifies DNA methyltransferase activity. In addition, we show that HP1β, a known SUV39H1-interacting protein, can bind directly to Dnmt1 as well as Dnmt3a and that endogenous HP1β purifies DNA methyltransferase activity. Taken together, the present study reveals a direct link between the enzymatic activities responsible for two distinct epigenetic layers.
Recently, a connection between DNA and histone methylation has been uncovered in N.crassa and A.thaliana. It was found that the genes dim-5 in N.crassa and kryptonite in A.thaliana encode H3-K9-specific HMTase. Interestingly, mutations in dim-5 or kryptonite that abolish their enzymatic activity lead to a decrease in DNA methylation (11,12). The mechanisms by which histone methylation influences DNA methylation are still unclear. Given that in A.thaliana the CMT3 DNA methyltransferase associates with the Arabidopsis homologue of HP1, LHP1, it was proposed that Kryptonite would methylate H3-K9, creating a binding platform for LHP1. CMT3 would then be recruited by the adaptor LHP1 to methylate DNA and thereby DNA methylation would be dependent on prior histone methylation (12). Our data suggest that a similar scenario could also be operational in mammals. We found that endogenous HP1β purifies DNA methyltransferase activity and, in agreement with our observation, Dnmt3a was found to co-localise with HP1β (6). Thus, similarly to what was found in Neurospora and Arabidopsis, mammalian Dnmts could be recruited by the adaptor molecule HP1 to chromatin that contains methylated histones. In addition, our finding that the DNA methyltransferases themselves associate with the H3-K9 SUV39H1 HMTase extend this initial model and imply a much more direct connection between DNA methylation and histone methylation. Indeed, the present data suggest that the Dnmts could be directly recruited to H3-K9 methylated chromatin through their interaction with the SUV39H1 histone methyltransferase. This possibility is interesting because it would mean that the histone methyltransferase activity associated with the DNA methyltransferases may be required for the Dnmts to methylate DNA. In other words, the Dnmts could only access and methylate DNA that is first wrapped around histones modified at Lys9 of H3. The notion that chromatin remodelling is required for DNA methylation to occur is supported by recent studies on the SNF2-like ATPase family. In Arabidopsis, mutation of the putative SNF2-like ATPase DDM1 leads to a severe reduction in DNA methylation (24) and targeted disruption of its mammalian homologue LSH causes loss of DNA methylation and lethality in mice (25). These data suggest that the putative nucleosome remodelling activity of these ATPases is required so that DNA becomes suitable to serve as a substrate for methylation.
A model of epigenetic transmission in the opposite direction should also be considered, namely that DNA methylation can influence the methylation of histones. This reverse scenario is supported by our recent finding that the methyl-CpG-binding protein MeCP2 associates with H3-K9 methyltransferase activity and is involved in targeting methylation of histone H3 Lys9 to a DNA methylated gene that it regulates (13).
Taken together, the recent findings in Neurospora and Arabidopsis as well as our data suggest the following order of events that connect DNA to histone methylation. In the initial phase, the Dnmts would add methyl groups to DNA only on chromatin that is methylated at Lys9 and bind HP1. The direct physical link identified here between the Dnmts and the SUV39H1–HP1 system would make sure that the H3-K9 methylation status directly influences DNA methylation patterns. In a second step, the generation of methylated DNA by the Dnmts would allow DNA binding of MeCP2, which in turn associates and favours histone methylation at Lys9 of H3. This sequential process of coupling DNA with histone methylation is particularly attractive because it would suggest that DNA methylation may also feed back to facilitate histone methylation, thereby reinforcing the two modes of epigenetic silencing and creating a self-propagating epigenetic cycle for long-term transcriptional repression.
Beside contacting H3-K9 HMTase activity, we found that Dnmt3a also associates, albeit weakly, with histone methyltransferase activity at Lys4 of H3. This finding was somewhat unexpected as DNA methylation is associated with gene silencing and H3-K4 methylation has been described to correlate with transcriptional activation (19). However, recent work in Saccharomyces cerevisiae indicates that the Set1 H3-K4 HMTase is required for gene silencing of rDNA (26,27). The mechanisms by which H3-K4 methylation is linked to transcriptional silencing are currently unknown. One explanation could be related to the histone code hypothesis (28), namely that the effect of H3-K4 methylation on transcription may well depend on its combination with other histone modifications. Our finding that a DNA methyltransferase can contact H3-K4 activity opens up an alternative possibility whereby methylation at Lys4 of H3 might function in concert with DNA methylation to repress transcription. By analogy to what we propose here for the connection between the Dnmts and H3-K9 methylation, it is possible that H4-K4 HMTase recruits the Dnmts to the DNA that has to be modified or conversely DNA methyltransferases may influence H3-K4 methylation. Future work will be needed to test this attractive possibility.
In conclusion, our data further emphasise the intimate relationship that exists between DNA methylation and histone methylation. The physical interaction identified here between the enzymes responsible for these distinct methylation layers further substantiates the concept of a self-reinforcing cycle for the propagation of a stable state of inactive chromatin.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Li for the Dnmt3a antibodies (164a and 164b), R. Laskey for anti-HP1β antibody and A. Jeltsch for the pET-Dnmt1 constructs. We are grateful to Helena Santos-Rosa for advice on histone methyltransferase assays. F.F. was funded by the Belgian Fonds National de la Recherche Scientifique (‘Chargé de Recherches du FNRS’) and R.D. was supported by the Belgian FRIA. The research in the T.K. laboratory is supported by a programme grant from the Cancer Research Campaign.
REFERENCES
- 1.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed]
- 2.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed]
- 3.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed]
- 4.Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed]
- 5.Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces and chromatin. Cell, 99, 451–454. [DOI] [PubMed]
- 6.Bachman K.E., Rountree,M.R. and Baylin,S.B. (2001) Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem., 276, 32282–32287. [DOI] [PubMed]
- 7.Fuks F., Burgers,W.A., Brehm,A., Hughes-Davies,L. and Kouzarides,T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed]
- 8.Fuks F., Burgers,W.A., Godin,N., Kasai,M. and Kouzarides,T. (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J., 20, 2536–2544. [DOI] [PMC free article] [PubMed]
- 9.Robertson K.D., Ait-Si-Ali,S., Yokochi,T., Wade,P.A., Jones,P.L. and Wolffe,A.P. (2000) DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet., 25, 338–342. [DOI] [PubMed]
- 10.Rountree M.R., Bachman,K.E. and Baylin,S.B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet., 25, 269–277. [DOI] [PubMed]
- 11.Tamaru H. and Selker,E.U. (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature, 414, 277–283. [DOI] [PubMed]
- 12.Jackson J.P., Lindroth,A.M., Cao,X. and Jacobsen,S.E. (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 416, 556–560. [DOI] [PubMed]
- 13.Fuks F., Hurd,P.J., Wolf,D., Nan,X., Bird,A.P. and Kouzarides,T. (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem., 278, 4035–4040. [DOI] [PubMed]
- 14.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed]
- 15.Nielsen S.J., Schneider,R., Bauer,U.M., Bannister,A.J., Morrison,A., O’Carroll,D., Firestein,R., Cleary,M., Jenuwein,T., Herrera,R.E. and Kouzarides,T. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature, 412, 561–565. [DOI] [PubMed]
- 16.Fatemi M., Hermann,A., Pradhan,S. and Jeltsch,A. (2001) The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N‐terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol., 309, 1189–1199. [DOI] [PubMed]
- 17.Deplus R., Brenner,C., Burgers,W.A., Putmans,P., Kouzarides,T., de Launoit,Y. and Fuks F. (2002) Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res., 30, 3831–3838. [DOI] [PMC free article] [PubMed]
- 18.Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed]
- 19.Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed]
- 20.Aagaard L., Schmid,M., Warburton,P. and Jenuwein,T. (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci., 113, 817–829. [DOI] [PubMed]
- 21.Peters A.H., O’Carroll,D., Scherthan,H., Mechtler,K., Sauer,S., Schofer,C., Weipoltshammer,K., Pagani,M., Lachner,M., Kohlmaier,A., Opravil,S., Doyle,M., Sibilia,M. and Jenuwein,T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337. [DOI] [PubMed]
- 22.Aagaard L., Laible,G., Selenko,P., Schmid,M., Dorn,R., Schotta,G., Kuhfittig,S., Wolf,A., Lebersorger,A., Singh,P.B., Reuter,G. and Jenuwein,T. (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed]
- 23.Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed]
- 24.Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed]
- 25.Dennis K., Fan,T., Geiman,T., Yan,Q. and Muegge,K. (2001) Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev., 15, 2940–2944.11711429
- 26.Briggs S.D., Bryk,M., Strahl,B.D., Cheung,W.L., Davie,J.K., Dent,S.Y., Winston,F. and Allis,C.D. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev., 15, 3286–3295. [DOI] [PMC free article] [PubMed]
- 27.Bryk M., Briggs,S.D., Strahl,B.D., Curcio,M.J., Allis,C.D. and Winston,F. (2002) Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol., 12, 165–170. [DOI] [PubMed]
- 28.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed]