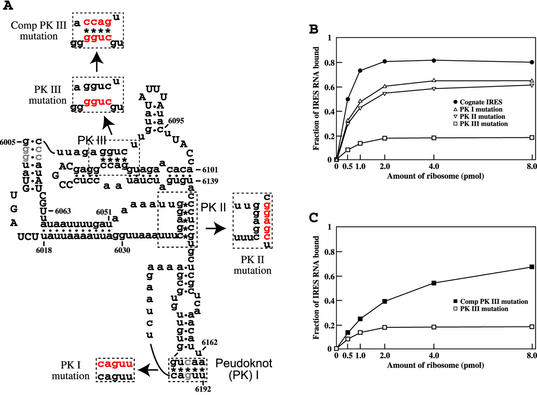
Figure 1.
Effect of base pairing in IRES PKs on 80S ribosome binding. (A) Secondary structure model of PSIV IRES RNA. Asterisks and dots indicate base pairing in PKs and helical stems, respectively. Highly conserved nucleotides in CrP-like viruses are capitalized. Half-tones denote nucleotides mutated in these studies to produce the cognate IRES (see Materials and Methods). Nucleotides mutated in PKs, to disrupt base pairing and to restore base pairing by compensatory mutations, are in red. (B) Binding of 32P-labeled IRES RNAs to 80S ribosomes. The fraction of IRES RNA bound is the ratio of [32P]RNA retained on the filter to that of the input [32P]RNA. (C) Effect of the compensatory mutations restoring base pairing in PK III on ribosome binding.