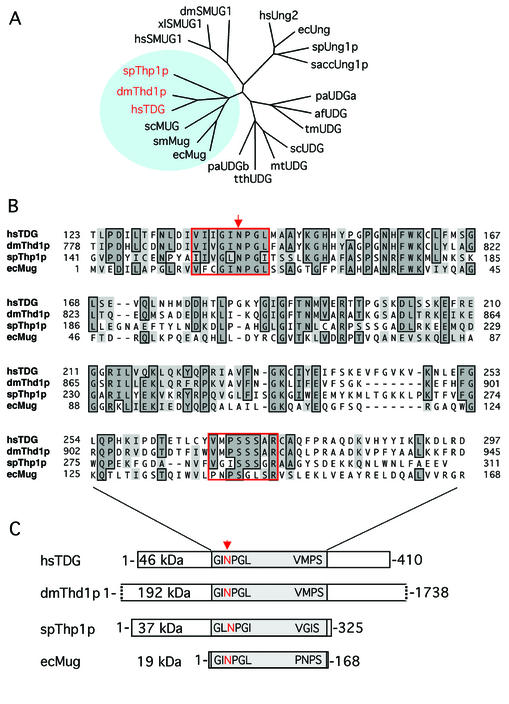
Figure 1.
Conservation of MUG-type UDGs. (A) Shown are the phylogenetic relationships between representative UDGs of the α/β-fold superfamily. Included are Homo sapiens UNG2 (hsUNG2; accession no. P22674), TDG (hsTDG; Q13569) and SMUG1 (hsSMUG1; O95862); Drosophila melanogaster Thd1p (dmThd1p; Q9V4D8) and SMUG1 (dmSMUG1; Swiss-Prot, Q9VEM1); Xenopus laevis SMUG1 (xlSMUG1; Q9YGN6) Schizosaccharomyces pombe Ung1p (spUng1p; O74834) and Thp1p (spThp1p: O59825); Saccharomyces cerevisiae Ung1p (saccUng1p; P12887); Serratia marcescens Mug (smMug; P43343); Escherichia coli Ung (ecUng; P12295) and Mug (ecMug; P43342); Streptomyces coelicolor UDGb (scUDGb; NP_626251) and MUG (scMug; NP_625542); Pyrobaculum aerophilum UDGa (paUDGa; NP_558739) and UDGb (paUDGb: NP_559226); Thermus thermophilus UDG (tthUDG; CAD29337); Mycobacterium tuberculosis UDG (mtUDG; NP_335742); Thermotoga maritima UDG (tmUDG; NP_228321); and Archaeoglobus fulgidus UDG (afUDG; NP_071102). (B) Amino acid sequence alignment of the catalytic core domains of spThp1p, dmThd1p with hsTDG and ecMug. The conserved MUG characteristic active site motif G(I/L)NPG(L/I) in the N‐terminal part of the catalytic domain and the less conserved C-terminal residues that are critical for a specific interaction with the substrate DNA are framed in red; other conserved residues are framed and shaded. The arrow indicates the position of the critical catalytic residue (N140) of TDG (6). (C) Schematic representation of the overall domain organization of the same members of the MUG subfamily of UDGs. The conserved core domains of the proteins are shaded, and the relative positions of the active site motifs indicated. All sequence analyses were done with the ClustalW routine (Blosum30 matrix; Gap penalty, 10; Gap extension penalty, 0.1) of the MacVector Sequence analysis software (Version 7.1.1; Accelrys, Burlington, USA).