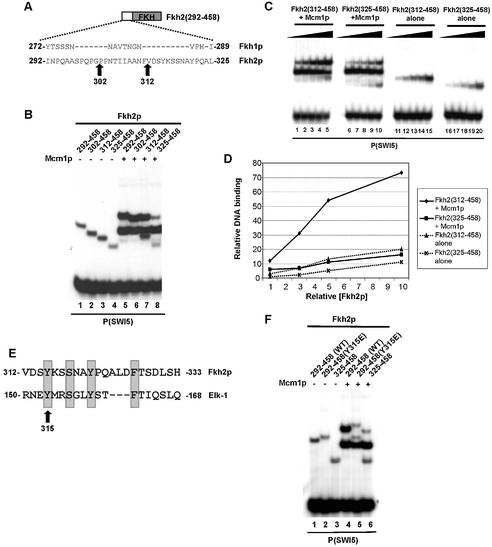
Figure 3.
Fine mapping of the Fkh2p ‘cooperativity domain’. (A) A schematic of the truncated Fkh2(292–458) protein. The amino acid sequence of the sequence immediately preceding the FKH domain of Fkh2p and the analogous region in Fkh1p are shown. The amino acids at the truncation endpoints are highlighted. (B) Gel retardation analysis of the indicated Fkh2p truncated proteins in the absence (lanes 1–4) or presence (lanes 5–8) of Mcm1(1–96) on the P(SWI5) site. (C) Gel retardation analysis of increasing amounts of the truncated Fkh2p truncated proteins in the presence (lanes 1–10) or absence (lanes 11–20) of Mcm1(1–96). Relative molar amounts of Fkh2p were 1 (lanes 1, 6, 11 and 16); 3 (lanes 2, 7, 12, 17); 5 (lanes 3, 8, 13, 18); 10 (lanes 4, 9, 14, 19); 20 (lanes 5, 10, 15, 20). (D) Quantification of the data in (C). The binding observed with the highest concentrations of Fkh2p used is not shown as the binding in lane 5 is beyond the linear range. (E) Alignment of amino acids 312–333 of Fkh2p with the B-box region (amino acids 150–168) of Elk-1. Amino acids in Elk-1 that affect interactions with SRF by >50% (18) are shaded. These are conserved in Fkh2p. (F) Gel retardation analysis of the indicated Fkh2p mutant proteins in the absence (lanes 1–3) or presence (lanes 4–6) of Mcm1(1–96) on the P(SWI5) site.