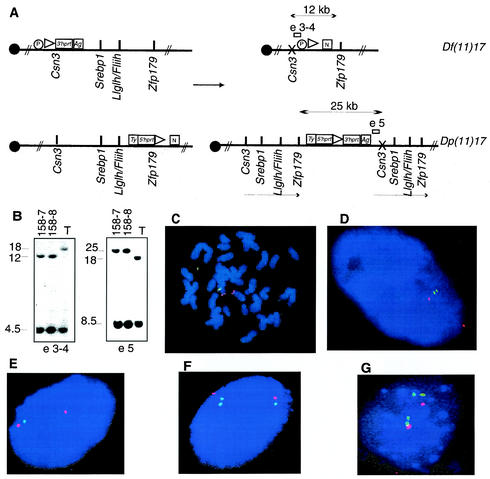
FIG. 2.
Genetically engineered chromosomes. (A) Double-targeted ES cell clones were subjected to transient expression of Cre recombinase. The predicted products of the recombination are schematized, with arrows indicating the expected fragment sizes when genomic DNA from rearranged chromosomes was digested with NdeI. Open rectangles represent the probes used in genomic Southern analysis of ES cells; probe e3-4 is predicted to detect a 12-kb band in the cell line with the deletion, whereas e5 is anticipated to detect a 25-kb band in the cell line with the duplication. The presence of an X over Csn3 denotes that a mutation of this gene is achieved by the insertion of the targeting vector. Ty, tyrosinase minigene; Ag, K14Agouti transgene; N, neomycin resistance gene; P, puromycin resistance gene. (B) Southern analysis of ES clones selected by using HAT media after the induction of Cre-loxP-mediated recombination. Two clones, 158-7 and 158-8, were analyzed sequentially with both probes, and the expected pattern for duplication and deletion of the predicted regions was identified in genomic DNA digested with NdeI. T, double-targeted ES cell not subjected to Cre recombinase. Independent confirmation of the predicted structure for rearranged chromosomes was obtained by FISH analysis with large insert BAC probes. (C) ES cells from clone 158-7 with deletion and duplication metaphase chromosomes were analyzed with probes containing Fliih (green) as well as D11Mit11 (red) as a reference probe that lies outside the rearranged interval. Note the absence of a green (Fliih) signal on one chromosome 11 homologue, signifying deletion. (D) Interphase nuclei from the same ES cell clone probed with Srebp1 (green) and Pmp22 (red) probes. The extended interphase chromosomes enable visualization of the submicroscopic duplicated interval (two nearby green dots). Identical results were obtained with ES cell clone 158-8. (E, F, and G) Depicted are results from FISH analysis of interphase nuclei of tail fibroblasts obtained from animals with the following genotypes: Df(11)17/+, Dp(11)17/+, and Dp(11)17/Dp(11)17, respectively. Srebp1 (green) and Pmp22 (red) were used as probes.