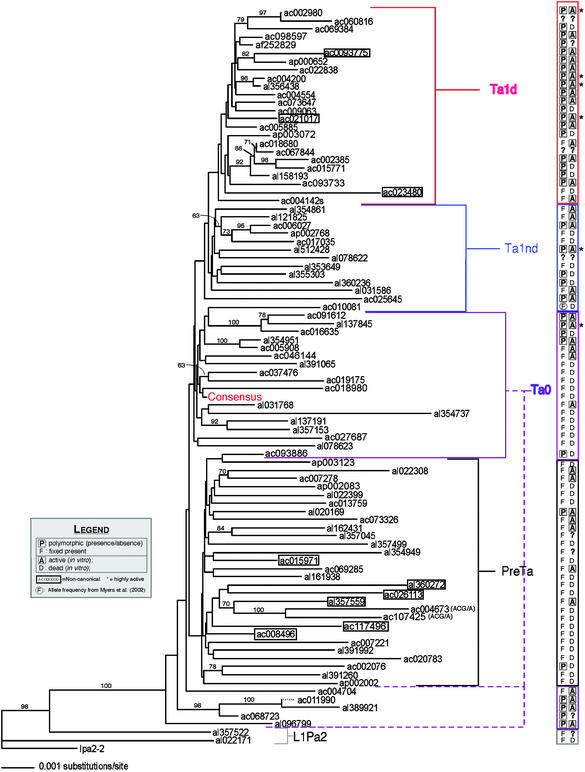
Figure 3.
Neighbor-joining tree with 89 intact L1 elements. The tree was constructed by using the full L1 sequences as described in Materials and Methods. The nodes recovered >60% of the time in 1,000 bootstrap replicates of the data are indicated, excluding CpG dinucleotides and the polypurine tract in the 3′ UTR. Ac009269 with the ≈400-bp deletion in the 5′ UTR was not included. Polymorphism and activity data are appended in the column on the right. A question mark signifies that the experiment was not performed. Hot elements are followed by asterisks, and noncanonical elements are boxed. The consensus sequence of the 89 elements is indicated. The Ta-0 subgroup that clusters between the ancestral L1Pa2 elements and the pre-Ta group had been identified (31). Based on short branch lengths, polymorphism data, and the in vivo activity of one member (LRE2/al389921) (21), the subgroup was predicted to be relatively young (31). We verify that prediction by showing four of five tested members to be polymorphic and all tested members to be active. This group is canonically Ta-0 based on seven defining nucleotides (Table 1). However, the group appears similar to L1Pa2 ancestral L1s based on its position in the tree. Preliminary sequence analysis comparing these L1s to the two L1Pa2 elements shows ancestral nucleotides spread throughout each element in a mosaic pattern and no obvious region where an element is clearly ancestral or young. AC004673 and AC107425 are members of the ACG/A group, distinct from the pre-Ta subgroup. They are marked as such.