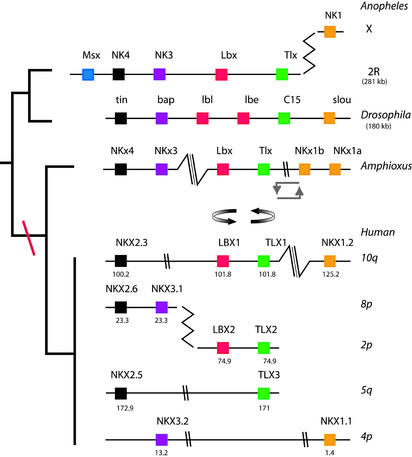
Figure 4.
Comparison of NK homeobox gene clusters between Anopheles, Drosophila, amphioxus, and human chromosomes. Boxes indicate homeobox genes, color coding denotes orthologous relationships. Cluster breaks are denoted by double-parallel marks (when intergenic distance >1 Mb), triple parallel marks (very large distances), or zigzag (transposition between chromosomes). Angular double arrows denote a large chromosomal inversion in amphioxus; curved double arrows denote a local inversion of a gene pair (relative to the Drosophila gene order). Genomic location is shown for human.