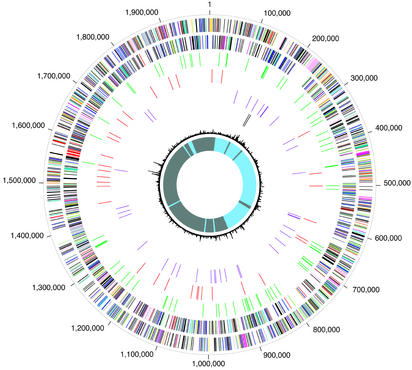
Figure 2.
Circular representation of the C. burnetii RSA493 overall genome structure. The outer scale designates coordinates in base pairs. The first circle shows predicted coding regions on the plus strand color-coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows predicted coding regions on the minus strand color-coded by role categories: third circle, pseudogenes in green; fourth circle, IS elements in red; fifth circle, tRNA genes in violet; sixth circle, rRNA genes in dark brown; seventh circle, trinucleotide composition in black; eighth circle, GC-skew curve in blue (positive residues) and gray (negative residues).