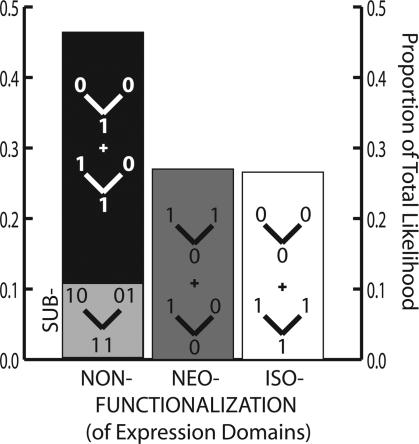
Fig. 3.
We examined the likelihood that observed patterns of gene expression were generated by three different processes, non-, neo-, or iso-functionalization of gene expression domains. We considered every node in every three-gene phylogeny and every expression domain of Fig. 1 and summed likelihood values over all possible ancestral states. Consistent with Fig. 2A, nonfunctionalization (repression) is likely to account for more of the observed patterns than neofunctionalization (activation). A subset of the likelihood attributable to domain nonfunctionalization (note we refer here to loss of expression, not loss of an entire gene, as usually implied by the term nonfunctionalization) is consistent with a mechanism of complementary repression (subfunctionalization), leading to specialization of ancestral gene function.