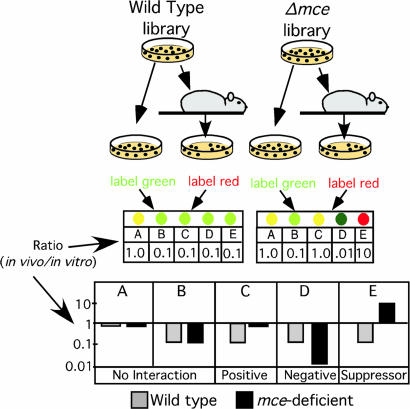
Fig. 1.
Strategy for identifying genes that interact with the mce1 and mce4 loci. Saturated transposon libraries were generated in wild-type or mce-deficient strains and the resulting libraries were used to infect groups of mice. After a period of infection, the surviving bacteria were recovered from spleen homogenates by plating. The unselected pools were replated in parallel to serve as a control. Each of these libraries was collected and “TraSH probe” was generated, which was complementary to the chromosomal DNA flanking each insertion in a pool. In vitro- and in vivo-selected pools were labeled with different fluorophores and compared by competitive hybridization to a microarray. (Lower) Idealized data. Genes (A–E) that are specifically required for in vivo growth will produce microarray ratios (in vivo/in vitro) of less than 1. Transposon mutations that cause a quantitatively similar phenotype in both genetic backgrounds (i.e., no interaction) will produce the same microarray ratios in both pools (gene B). Positively interacting transposon mutations will produce numerically larger ratios in the mutant background (gene C), and negatively interacting mutations will produce numerically smaller ratios (gene D). Suppressor mutations will produce ratios greater than 1 in the mutant background (gene E).