Abstract
The free energy landscape for folding of the Alzheimer's amyloid-β(25–35) peptide is explored using replica exchange molecular dynamics in both pure water and in HFIP/water cosolvent. This amphiphilic peptide is a natural by-product of the Alzheimer's amyloid-β(1–40) peptide and retains the toxicity of its full-length counterpart as well as the ability to aggregate into β-sheet-rich fibrils. Our simulations reveal that the peptide preferentially populates a helical structure in apolar organic solvent, while in pure water, the peptide adopts collapsed coil conformations and to a lesser extent β-hairpin conformations. The β-hairpin is characterized by a type II′ β-turn involving residues G29 and A30 and two short β-strands involving residues N27, K28, I31, and I32. The hairpin is stabilized by backbone hydrogen-bonding interactions between residues K28 and I31; S26 and G33; and by side-chain-to-side-chain interactions between N27 and I32. Implications regarding the mechanism of aggregation of this peptide into fibrils and the role of the environment in modulating secondary structure are discussed.
INTRODUCTION
Alzheimer's disease is a neurological disorder associated with the pathological self-assembly of the Alzheimer amyloid-β (Aβ) peptide into toxic soluble oligomers and insoluble fibrils with high β-sheet content. The Aβ peptides are proteolytic by-products of the transmembrane amyloid precursor protein (APP) (1). As part of the APP complex, the Aβ peptide's hydrophilic N-terminus is exposed to the aqueous extracellular environment while its hydrophobic C terminus is embedded in the membrane (2). Upon proteolytic cleavage, the peptide is released into the extracellular milieu, where, under appropriate cellular conditions, aggregation can occur. The predominant forms of the Aβ peptide present in aggregates are 40–42-amino-acids long, although other lengths can be present as well. In particular, an 11-residue-long fragment, Aβ(25–35) (with sequence GSNKGAIIGLM), is produced in the brains of aged patients from proteolytic cleavage of soluble racemized Aβ(1–40) peptides (3). The Aβ(25–35) peptide possesses many of the characteristics of the full-length Aβ(1–40/42) peptide, including an amphiphilic nature and an ability to aggregate, but its small size renders it a more attractive model system to study the conformational changes involved in Alzheimer's disease. Aggregates of Aβ(25–35) have been shown to possess the neurotrophic and neurotoxic properties of their full-length counterparts (4,5) and there is evidence that the monomeric form of the peptide may itself be cytotoxic (6).
Unlike proteins, most peptides do not possess a unique, stable, well-defined three-dimensional structure, but populate a variety of partially structured or even completely unfolded conformations under physiological conditions. The diversity of conformations that peptides can adopt renders the study of these peptides a challenge, particularly from an experimental standpoint. Aβ peptides in particular do not lend themselves to structural experimental characterizations in aqueous solvent, as they have low solubility and aggregate under the concentration conditions (typically >1 mM) required for NMR studies. As a result, the three-dimensional structure of Aβ(25–35) peptide in water is not known. It is, however, critical to determine the monomeric conformations of this peptide as they can play an important role in determining the nature of early aggregates and the resulting morphology of the fibril. Indeed, different monomer conformations can lead to different intermediate species for aggregation, and eventually to fibrils of different shapes. Evidence that different initial seeds lead to different fibrils is given in the recent work by Petkova et al. (7), in which different sample preparation schemes resulted in altered fibril morphologies.
Because of the inherent difficulties associated with working on the Aβ(25–35) peptide in water, effort geared at characterizing the three-dimensional structure of this peptide have thus far been limited to NMR studies performed either in water/organic solvent mixtures or in micellar solutions (8,9). The secondary structure has been probed through circular dichroism (CD), vibrational circular dichroism, and Fourier transform infrared spectroscopy (10–14) in a variety of solvents. The conformations adopted by the peptide are extremely sensitive to the media involved. As a general rule, the peptide adopts a helical structure in apolar organic solvents (such as TFE and HFIP) and an unstructured conformation or β-structure (β-turn or β-sheet) in aqueous buffer or polar organic solvent (8,9,13), although these trends can be altered by pH, concentration, incubation time, and the preparation and purification process (10–15). An atomically detailed characterization of the structures adopted by Aβ(25–35) in water has yet to emerge from experimental studies.
In this work, we use replica exchange molecular dynamics (REMD) simulations to probe the structure of the Aβ(25–35) peptide in both a membrane-mimicking environment (HFIP/water) and in the extracellular environment (pure water). An advantage of simulations over traditional bulk experiments lies in treating the peptide at a single-molecule level, hence permitting the identification of structured conformations of low population that would not be seen in ensemble measurements. Such conformations may play a critical role in initiating aggregation and in determining the morphology of resulting fibrils. Our first simulations, in HFIP/water cosolvent, allow us to make a direct comparison with experiment. We find, in agreement with experiment, that the peptide adopts a primarily helical structure in apolar organic solvent. We then turn to a prediction of the conformations sampled in aqueous solvent and find that the peptide adopts vastly different structures than in the HFIP/water mixture, now coexisting between β-hairpin and collapsed-coil configurations. The nature of these structures, as well as their possible role in initiating aggregation, will be discussed.
MATERIALS AND METHODS
Simulation methodology
All the simulations were started from the minimized solution NMR structure (PDB code: 1QWP) of the Aβ(25–35) peptide in 80:20 (vol/vol) HFIP/water mixture. The NMR structure consists of an α-helix involving residues 28–31 and a 310-helix involving residues 32–34. The peptide was solvated in water and in a 80:20 HFIP/water mixture. Water was modeled by the explicit simple point-charge model (16) and HFIP by the all-atom model developed by Fioroni et al. (17). The simulations were performed using periodic boundary conditions in a dodecahedron box, with the minimum distance between the solute and the box wall being 1.4 nm.
The MD simulations were performed using the GROMACS software package (18,19) and GROMOS96 force field (20). The LINCS algorithm was used to constrain all bond lengths in the peptides and HFIP and the SETTLE algorithm for the water molecules, allowing an integration time step of 2 fs. A twin-range cutoff 0.9/1.4 nm was used for the nonbonded interactions, and a reaction-field correction with dielectric permittivity ε = 80.1(30.3) (17) was employed to calculate long-range electrostatics interactions in pure water (80:20 HFIP/water). The temperature and the pressure were maintained by coupling temperature and pressure baths using the method of Berendsen et al. (21). The solute and solvent were separately coupled to external temperature and pressure baths. The temperature-coupling constant was 0.1 ps. The pressure was kept at 1 bar using weak pressure coupling with τp = 1.0 ps (21).
The system was energy-minimized by steepest descent for 1500 steps. In all the simulations the solvent was equilibrated in a 100-ps MD run with position restraints on the peptide. The solvent equilibration run was followed by another 100-ps run without position restraints on the peptide. The density of the solvent for water+peptide (WP) and for HFIP/water+peptide (HWP) system is 975 kg/m3 and 1445 kg/m3, respectively. After equilibration, two different REMD simulations (NVT ensemble for each replica) were performed for the WP and HWP systems. REMD is an enhanced sampling protocol (22,23), in which several identical copies (replicas) of the system are run in parallel at different temperatures and are periodically swapped with a probability given by the Metropolis criterion (24–27). This leads to escape from low-lying energy traps and enhanced equilibration.
For the HWP system, two 16-ns independent REMD simulations were performed. Each REMD run consists of 34 replicas, and the total simulation time is 544 ns for each REMD run. The temperature ranges from 270 K to 485 K with exponential distribution. The swap time between neighboring replicas is 3 ps. The acceptance ratio is between 20.1% and 27.7%. In addition, two independent 20-ns standard molecular dynamics simulations at 300 K and 1 bar were performed for the peptide. For the WP system, two 16-ns independent REMD simulations were performed. Each REMD run consisted of 40 replicas and the total simulation time is 640 ns for each REMD run. The temperature ranges from 300 K to 515 K with exponential distribution. The swap time between neighboring replicas is 2 ps and acceptance ratio ranged from 16.5% and 30.2%.
Analysis methods
Trajectory analyses were performed with the facilities implemented in the GROMACS software package (18,19). The secondary structure content was identified using the DSSP program (28). A hydrogen bond (H-bond) is considered formed if the donor-acceptor distance is <0.35 nm and the donor-hydrogen-acceptor angle >120°. A helical structure is present if at least three (four or five) consecutive residues have helical structure content (including 310-helix, α-helix, and 5-helix). A β-hairpin conformation exists if at least four residues have a β-sheet conformation and two residues have a β-turn conformation. A side-chain-to-side-chain interaction exists if the distance between the center of mass of the side chains of two residues is smaller than 0.60 nm. The Daura cluster analysis method (29) was used to cluster the conformations sampled in the REMD simulations. In a first step, the peptide backbone (the two residues in the N- and C-termini were not considered due to their high flexibility) root mean-square deviation (RMSD) between all pairs of structures was calculated. The structure with the largest number of neighbors satisfying the condition that the RMSD from the central structure of the cluster is ≤0.1 nm was taken, together with the neighbors, to form the first cluster and eliminated from the pool of structures. This process was repeated until no structures remained in the pool.
The VMD and MOLMOL programs were used for trajectory visualization and for graphical structure analysis. All simulations were performed on 128 dual CPU Xeon 3.06 GHz processors. In all the trajectory analysis for REMD runs, the data generated during the first 2 ns were not included, and all the results are an average of two independent 16-ns REMD runs, except when mentioned otherwise. End-to-end distance (dd) used in this study is the distance between the α-carbon atom of S26 and that of L34. RMSD is the backbone RMSD for residues 26–34 of the Aβ(25–35) peptide except when mentioned otherwise.
RESULTS
Conformational states of Aβ(25–35) in HFIP/water cosolvent: stabilization of helical structure
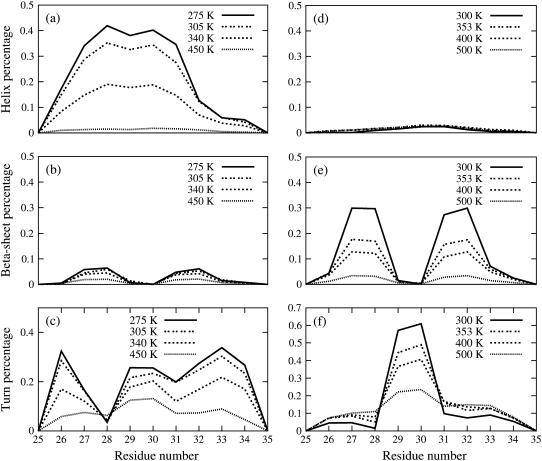
The conformations sampled during the two REMD runs were clustered by mutual RMSD as described in Materials and Methods. At 275 K, the peptide is seen to exist 45% of the time in a helical conformation. The helical structures coexist with unstructured coils of similar dimensions to the helix, as well as, to a much lesser extent, compact, collapsed coils of smaller dimensions than the helix. The secondary structure probability per residue is shown at four different temperatures in Fig. 1. The helical structures possess a stable central α-helix (residues 27–31), while the terminal residues show more fluctuations. The C-terminus adopts a mostly turnlike structure, while the N-terminal region alternates between a turn and a helical structure. Collapsed coils generally possess turns in the central region of the peptide (residues 29–30). The helical structures found in our simulations in HFIP/water are in good agreement with the structure derived from NMR. NMR studies show that Aβ(25–35) adopts a turn-helical structure in solutions containing at least 50% HFIP in volume. This structure has a partially ordered turn in the N-terminus (residues 26–28), followed by an α-helix from residues 28–31, and a 310-helix spanning the C-terminus residues 32–34. Our simulations indicate (Fig. 1) that the helix vanishes at high temperature (450 K), while the turn structures appear to be more resistant to temperature denaturation.
FIGURE 1.
The percentage of helical, β-sheet, and turn conformation for each residue of Aβ(25–35) at four different temperatures (300 K, 353 K, 400 K, and 500 K) in HFIP/water mixture (a–c) and in pure water (d–f).
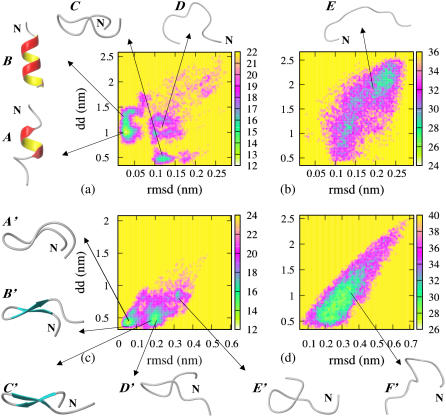
The potential of mean force (PMF) for the peptide in HFIP/water cosolvent is given as a function of end-to-end distance (dd) and backbone RMSD from the central 28–31 residues (α-helix) in the NMR PDB structure at 275 K and 450 K in Fig. 2, a and b. The backbone RMSD from the central-positioned four residues (28–31) was chosen as a reaction coordinate as this segment forms a stable α-helix (while the terminal residues show significant structural fluctuations). Three minimum-energy basins were found in the PMF plot at 275 K (Fig. 2 a). They are located at (RMSD, dd) values of (0.025 nm, 1.0 nm) (the lowest minimum-energy basin), (0.125 nm, 0.45 nm), and (0.12 nm, 1.1 nm), corresponding to helical structure, a β-hairpin-like collapsed coil with turn in the central region (G29-A30), and a more extended coil conformation, respectively. At 450 K, the peptide has lost all helicity and samples different extended coil conformations (Fig. 2 b). Representative structures located in the different minimum-energy basins are shown beside the PMF plot, labeled from A ∼ E.
FIGURE 2.
Potential of mean force of Aβ(25–35). (a) HFIP/water mixture at 275 K and (b) at 450 K plotted as a function of end-to-end distance (dd) and RMSD (residues 28–31) of each conformation relative to the starting NMR structure (residues 28–31); (c) in water at 300 K and (d) at 500 K, plotted as a function of end-to-end distance (dd) and RMSD of each conformation relative to the structure of the central member of the most populated cluster (collapsed coil). Representative structures of Aβ(25–35) located in the different minimum-energy basins are shown beside the PMF plot. The N-terminus of the peptide is labeled with the letter N.
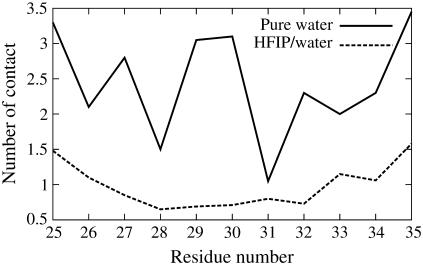
The role of HFIP/water mixture in stabilizing the helical structure was probed by investigating the interactions of the solvent molecules with the peptide backbone. Jasanoff and Fersht (30) have suggested that alcohols affect protein structure, in part, by displacing water molecules from hydrogen-bonding sites along the peptide backbone. Recent experiments and computer simulations indicate that preferential solvation on the surface of a peptide by fluoroalcohol component of a fluoroalcohol/water mixture may play an important role on the conformational effects of the peptide induced by fluoroalcohols (17,31–33). To test if preferential interaction between HFIP and peptide takes place for Aβ(25–35) in HFIP/water mixture, we considered two independent 20-ns-long molecular dynamics simulation at 300 K initiated from the NMR structure. During the course of one run, the amount of helicity fluctuated and we calculated the average number of contacts of both water molecules and HFIP molecules with the peptide at different points in the simulation. The average number of contacts of water molecules with the peptide in the initial 0.5th–1.5th-ns stages of the simulation (where helicity was moderate) and the final 1 ns of the simulations (where helicity was high) was monitored. The number of contacts of the water molecules with the peptide decreased by 32% from 131 to 89, while the number of contacts between the HFIP molecules and the peptide increased from 136 to 142. In addition, the average number of H-bonds between the water molecules and the peptide decreases from 14 and 10 during this same period, along with a decrease of peptide-water interaction (the corresponding interaction energy increases from −600 kJ/mol to −400 kJ/mol). Another 20-ns molecular dynamics run shows qualitatively the same results. These results support a picture in which the HFIP molecules displace the water molecules from the vicinity of the peptide, which in turn decreases the chance of forming H-bonds between either the peptide backbone and water or between amino-acid side chains and water. The large HFIP molecules coat the peptide, stabilizing an extended conformation and favoring the formation of helical intrapeptide H-bonds over β-sheet H-bonds. The hydration of the peptide by water was further investigated by analyzing the number of contacts between the water oxygen and the hydrogen-bond-forming group (−C=O and −N-H) of each residue in the first hydration shell (0.36 nm) during the duration of the REMD simulations (Fig. 3). In HFIP/water, residues located in the center of the peptide (the most helical region) have, on average, fewer hydration waters than those located near the N- and C-termini (where there is less structure). This shielding of the backbone is consistent with theoretical work by Vila et al. (34) and Ghosh et al. (35), in which peptide backbone dehydration is shown to stabilize helical conformations.
FIGURE 3.
Number of contacts between the oxygen of water and the backbone hydrogen-bond-forming groups (−C=O and −N-H) for each residue of Aβ(25–35) in the first solvation shell (within 0.36 nm) in HFIP/water mixture and in pure water.
Conformational states of Aβ(25–35) in water: adoption of collapsed-coil and β-hairpin structure
Having now established that the force-field and simulation methodology used for Aβ(25–35) correctly yield a helical conformation as the most stable structured state in HFIP/water, we turn to the prediction of the structure of Aβ(25–35) in water, using the same protein force field and the REMD approach.
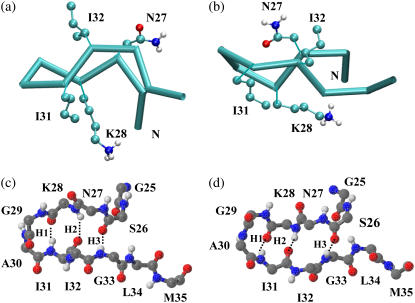
Structures obtained from the REMD simulations in water are clustered as described in Materials and Methods. We find that the conformational states populated by this peptide in pure water are in sharp contrast to those populated in HFIP/water cosolvent. The secondary structure probability per residue is shown in Fig. 1. The percentage of helical structure is insignificant (<3%) and the most populated conformation at 300 K is a collapsed coil conformation (populated 70% of the time). The collapsed coils coexist with β-hairpin conformations of similar dimensions (populated ∼30% of the time). Extended coils are rarely populated. The peptide adopts two different β-hairpin structures, shown in Fig. 4. Both structures are characterized by a type II′ β-turn involving residues G29 and A30, and two short antiparallel β-strands consisting of residues N27, K28, I31, and I32. The two types of β-hairpins differ in the twist of one strand relative to the other and have a mutual RMSD of 0.25 nm. The collapsed coils are of similar overall dimensions to the β-hairpin and have some structure present in the form of turns in the central region (30% of the time) as well as in smaller amounts in the N- (residues 26–27) and C-termini (residues 32–34). The temperature-dependence of the secondary structure is given in Fig. 1, d–f. While the population of β-sheets diminished significantly with increasing temperature, the turn structure located at G29 and A30 is thermally stable and retains a population of >20% at 500 K.
FIGURE 4.
Representative structures of the two most populated β-hairpin conformations. Both possess a type II′ β-turn involving residues G29 and A30 and two short β-strands involving residues N27, K28, I31, and I32, but differ in the relative twist of the strands. A trace representation of the β-hairpins and a CPK representation of the side chains of the β-stranded residues are shown in panels a and b. Structures a and b are located at positions (0.42 nm, 0.03 nm) and (0.44 nm, 0.19 nm), respectively, in Fig. 2. A fully atomic representation of the main chain of the two β-hairpins is shown in panel c (the same conformation as shown in a) and panel d (the same conformation as shown in b). H-bonds are represented by dotted lines and are numbered from the turn to the tail of the β-hairpin (H1–H3). The two β-hairpins have the same H-bond pattern. The N-terminus of the peptide is labeled with N.
The PMF of the Aβ25–35 peptide in water is plotted in Fig. 2, c and d, at 300 K and at 500 K as a function of the end-to-end distance (dd) and the RMSD from the central member of the most populated cluster (the collapsed coil structure labeled as A′ in Fig. 2 c). Representative structures belonging to each basin are labeled from A′∼ F′ in Fig. 2. At 300 K, there are three main basins located at (RMSD, dd) values of (0.05 nm, 0.45 nm), (0.2 nm, 0.5 nm), and (0.32 nm, 0.85 nm). The first basin is populated by collapsed-coil conformations with a β-turn located at G29–A30 (Fig. 2 c) as well as β-hairpin conformations. These collapsed coils do not satisfy the DSSP criteria (of proper intrapeptide H-bonds formed and (φ, ψ) values of residues N27, K28, I31, and I32 in the β-region of Ramachandran plot (28)) to form a true β-hairpin, but have low RMSD from the structured β-hairpin conformations. The second basin is populated with similar structures with a turn at G29-A30, but with a different twist of the strands from those in the first basin (see structures B′ and C′ in Fig. 2). The third basin corresponds to collapsed coils without this turn (although these states can possess turns in the N- and C-termini). These structures have larger end-to-end distance than those populated in the first two basins, but they are nonetheless quite compact (with dd <1 nm). At 500 K, the peptide loses its β-sheets' secondary structure and increases in size, now populating both collapsed and more extended coil conformations (Fig. 2 d). As noted previously, the G29-A30 turn is still present 20% of the time at 500 K.
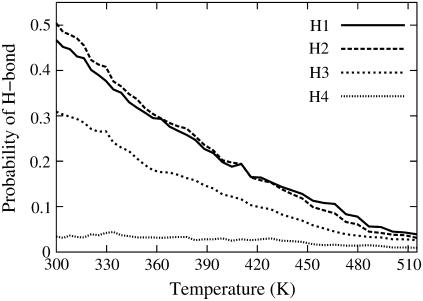
The interactions stabilizing the hairpin structure were probed by monitoring the probabilities of formation of four interstrand backbone hydrogen bonds: I31:HN-K28:O (H1), K28:HN-I31:O (H2), G33:HN-S26:O (H3), and S26:HN-G33:O (H4) (numbered from the turn to the tail of the β-hairpin) at a number of temperatures. The expression Xi:HN-Yj:O denotes a hydrogen bond between residues Xi and Yj, with X, Y being the one-letter code of amino acids and i, j being the sequence number in the Aβ(25–35) peptide. Fig. 5 shows the probability of each H-bond as a function of temperatures ranging from 300 to 515 K. H-bonds H1 and H2, located in the turn region of the β-hairpin, are seen to be much more stable than H-bonds H3 and H4, located at the extremity of the β-hairpin. H-bond H4 rarely formed and the β-hairpin structure appears to be mainly stabilized by H-bonds H1–H3. The probabilities of H-bonds H1–H3 decrease gradually with increasing temperature, consistent with the loss of hairpin structure seen in Fig. 1 e. Analysis of the probabilities of formation of four pairs of cross-strand side-chain-to-side-chain interactions: K28-I31, K28-I32, N27-I31, and N27-I32 at 300 K indicate that the dominant interaction in stabilizing the hairpin comes from the N27-I32 pair. This side-chain-to-side-chain contact is populated 60% of the time while the formation probabilities of the other three side-chain-to-side-chain interactions are <20%. Emerging from the interaction analysis is that the three interstrand H-bonds, H1–H3, and the side-chain-to-side-chain interaction between N27 and I32, play a significant role in the stabilization of the β-hairpin conformations sampled by Aβ(25–35) peptide in water.
FIGURE 5.
Probabilities of backbone H-bonds of Aβ(25–35) in water as a function of temperature.
An examination of the average number of contacts between water oxygen and the backbone hydrogen-bond-forming groups (−C=O and −N-H) for each residue in the first solvation shell (within 0.36 nm) at 300 K (Fig. 3) reveals that the backbone hydrogen-bond-forming groups of the β-turn residues G29 and A30 have higher number of contacts with water (∼3.0 for both of them) than the other residues of the peptide (not counting the solvent-exposed termini). This is in contrast to what we observed in cosolvent, where the central part of the peptide (now in helical form) shows less contact with water than the termini. The implication is that hydration/dehydration of the backbone affects the different conformational preferences of Aβ25–35 in the two different solvents and possibly their aggregation propensities. This will be discussed further in Discussion and Conclusions.
To check the force-field dependence on the main conformational states of Aβ(25–35) peptide adopted in water, a 32-ns REMD run with 40 replicas using OPLS/AA force field was performed. At 300 K, Aβ(25–35) is seen to adopt 2% helical structure and 23% of β-hairpin structure in OPLS/AA, in good agreement with the 3% of helical structure and 30% of β-hairpin structure found using the GROMOS96 force field. The main β-hairpin structure identified in the GROMOS simulations is also the predominant structure found in the OPLS/AA simulations. Slight variants of this β-hairpin (with different length of turn/bend conformations connecting the two β-strands) are also present in the OPLS/AA simulations, but they all involve residues G29 and A30 in the turn region. The length of the turn/bend varied from 2 ∼ 4 residues involving residues 28–31.
DISCUSSION AND CONCLUSIONS
Our simulations show that Aβ(25–35) adopts a mostly helical structure in HFIP/water cosolvent. In pure water, on the other hand, the peptide adopts mostly collapsed-coil structures as well as to a lesser extent β-hairpin conformations. The implication is that the Aβ(25–35) sequence has the intrinsic ability to populate a wide variety of secondary structures, ranging from helices to β-hairpins, and that solvents play the role of fine tuners of structure. Fluorinated alcohols such as HFIP and TFE are commonly used to stabilize helical structure, although in some instances, these solvents act as protein denaturants (36). Despite their frequent use, the precise mechanism by which they promote (or disrupt) structure is poorly understood. The use of REMD simulations enabled us to obtain, for the first time, a near-complete characterization of the free energy for folding of this peptide. Earlier theoretical work on small peptides in TFE and in HFIP were limited by computational resources, but nonetheless were able to suggest possible mechanisms for helix stabilization. Simulations by Brooks and Nilsson (36) on a blocked alanine tripeptide in TFE suggested that the fluoroalcohol adopted spatial and orientational order around extended conformations of the peptide. Fiorini et al. (17) studied the peptide melittin in a HFIP/water mixture in a five-nanosecond simulation and a 100-nanosecond simulation (37) initiated from a helical structure. Helicity was preserved during the simulation time, and HFIP molecules were seen to cluster around the peptide. Daidone et al. (38) have performed 50-ns-long simulations in TFE of the H1 prion peptide and of the 12–28 fragment of the Alzheimer amyloid-β peptide, with helical starting structures. Their simulations indicate that the helical forms of these peptides are very stable and are retained during the length of the simulation. Our simulations are consistent with previous simulations as well as recent NMR studies (33) and suggest that water is displaced from the immediate vicinity of the peptide, in favor of HFIP molecules. The latter arrange themselves around the peptide, preventing chain collapse and favoring the formation of helical intrapeptide hydrogen bonds over β-hairpin H-bonds.
Apolar solvents, such as a 80:20 (vol/vol) HFIP/water mixture, are useful probes of structure in a membranelike environment, but are not representative of the aqueous extracellular environment into which the peptide is released upon proteolytic cleavage. Mounting evidence point to the role of extracellular soluble species (monomers and oligomers), as well as fibrils as being the major players in cytotoxicity (39,40). Determining the monomeric structure of the peptide in aqueous solvent is a necessary first step to gain insight into possible modes of toxicity. While there are no NMR structures of Aβ(25–35) in pure water, CD measurements of a very dilute (50 μm sample of Aβ(25–35) peptide in freshly prepared phosphate buffer solution (PBS) suggest the presence of coil conformations, β-turn and β-sheet structures (41). These experimental conditions are believed to closely mimic aqueous conditions. Our theoretical findings, in which we find coexistence of collapsed coils and β-hairpins, are consistent with these experimental observations, and provide atomically detailed structural information about the conformations populated in aqueous solution. The hairpins are seen to possess a turn located in the A29-G30 region, and are stabilized by three H-bonds near the turn and by a side-chain-to-side-chain interaction involving residues N27 and I32. Two types of hairpins, differing in the relative twist of one strand with respect to the other, are observed. The coil structures adopt a variety of conformations, with turns in the central (A29-G30) region (the most common location), and in the N-terminus (26–28) and C-terminus (31–34). The coils structures have dimensions (as measured by the end-to-end distance) similar to those of the hairpin conformations (Fig. 2).
It is pertinent to compare the structure of the Aβ25–35 peptide in water studied here to that of other amyloidogenic fragments studied computationally. In particular, what is the nature of the structured states? Is the hairpin found in our simulations a common feature to amyloidogenic peptides? A hairpin must possess a minimum of four residues in order to form (42). Baumketner and Shea (43) have studied the monomeric and dimeric states of two amyloidogenic tetrapetides (KFFE and KVVE) using REMD simulations in implicit solvent. Their simulations indicate, however, that these peptides, in their monomeric form, do not adopt any hairpin conformations, but instead populate extended strandlike conformations and collapsed conformations lacking hairpin signatures. A slightly larger peptide, the seven-residue 16–22 fragment of the amyloid-β peptide was studied by Klimov and Thirumalai (44). Their eight-nanosecond simulation in explicit water indicated that the peptide existed in random coil and to a lesser extent in β-strand conformations. More recent work by Gnanakaran et al. (45) using REMD simulations in explicit solvent showed a dominant PPII conformation for this peptide. Larger amyloidogenic peptides, on the other hand, appear to populate β-hairpins. Replica exchange simulations of a peptide of similar length to the one studied here, the 11-residue fragment of β2 microglobulin, also showed the presence of β-hairpin conformations (46). The use of replica exchange (or other enhanced sampling techniques) is critical to achieve proper sampling of conformational space, particularly for larger peptides. Constant temperature simulations cannot reach timescales corresponding to the relaxation time of these peptides (on the order of milliseconds) and can therefore only provide a partial representation of the relevant conformations. Nonetheless, such constant temperature simulations have provided invaluable insight into some of the possible conformations adopted by amyloidogenic peptides. Constant temperature simulations (on the order of 100-ns-long) of the 14-residue-long H1-fragment of the prion protein and the 12–28 fragment of the amyloid-β protein (38) both showed population of hairpin structures in water. A microsecond-long simulation of the Alzheimer amyloid Aβ10–35 peptide identified the presence of a strand-loop-strand structure (47). REMD simulations in explicit solvent on the same peptide found that the peptide did not fold to a unique ground state, instead populating a number of collapsed coil conformations (A. Baumketner and J.-E. Shea, unpublished). Implicit solvent replica exchange simulations on the full-length Aβ1–42 peptide indicate that this peptide exists primarily as a collapsed coil, with small population of secondary structure (49). Constant temperature simulations of this peptide in explicit water, initiated from a helical structure found in apolar media, showed that one of the helices sampled β-rich and random coil structures, while the other remained helical (50).
A common feature to all these amyloidogenic peptides in aqueous solvent is that they exist mostly as collapsed coil states, populating only a fraction of structured states. This is not surprising, as one would expect such sequences to have frustrated landscapes, rather than the funneled landscape typical of well-designed proteins that fold to a unique global structure. Amyloidogenic peptides may have to populate a structured state with the correct geometry to self-replicate in order for aggregation to proceed. This could be a β-strand, in the case of shorter sequences, or a β-hairpin for longer sequences. We discuss next the possible role of the conformations found in our simulations with regard to aggregation.
The monomeric conformations in water identified in our simulations are all possible starting points for aggregation into fibrils. From an entropic standpoint, the most likely candidates to seed further growth are those with preformed structure (collapsed coils with turn formed, or β-hairpins). Indeed, these structures possess less configurational entropy than completely unstructured coil conformations, and will hence pay less of an entropic penalty for association into dimers and larger aggregates. Fernández and Scheraga (51) have noted that proteins that aggregate readily tend to have a significant number of backbone H-bonds exposed to solvent, available for further protein-protein interaction. Our simulations indicate (Fig. 3) that the backbone hydrogen-bond-forming groups of the β-turn region are more exposed to water than the remaining residues of the peptide. This suggests that the β-turn in Aβ(25–35) is a possible segment for initiating aggregation. (It is interesting to note that in cosolvent, where the peptide does not aggregate, all residues show low backbone H-bonds exposure to solvent.) A plausible scenario in which the turn region nucleates aggregation is through the formation of aggregates initiated from direct interpeptide interactions between the exposed H-bonds in the turn region. Two collapsed-coils conformations (possessing a turn in the G29-A30 region) would associate in a first step via their turn regions. This nucleation step would be followed by hydrogen-bonding and side-chain interactions between residues flanking the turn region, leading to aggregates formed of extended β-strands. Evidence supporting this scenario stems from CD measurements of Aβ(25–35) in PBS buffer solution (41). CD spectra of fresh samples showed the presence of β-turn, while 24 h samples (when aggregation is now well underway) show a net decrease in this motif, with the remaining monomeric structures now predominantly showing random coil (41) signatures. The implication is that the configurations possessing a turn structure have associated to form larger aggregates, showing their greater predisposition than the unstructured conformations to self-associate. Furthermore, the Aβ(25–35)(M35Nle) peptide, which does not aggregate readily, has considerably less β-turn content than the wild-type Aβ(25–35). In addition, the high thermal stability of the turn region found in simulation may enable this peptide to aggregate at high temperatures.
It may be noteworthy to compare the hairpin structure of Aβ(25–35), with its turn located at residues G29-A30, to proposed fibril structures of the full-length Aβ structure. Such comparisons must be taken with the caveat that it is not entirely clear whether the assembly of Alzheimer amyloid-β peptide fragments should proceed in the same way as does the full-length peptide. A fibril model by Petkova et al. (52), based on solid-state NMR, suggests the presence of a single loop involving a salt bridge between D23 and K28, while a recent model for quiescent Aβ(1–40) fibrils based on proline (and alanine) scanning mutagenesis data (53,54) suggest the presence of two turns located at positions E22-D23 and G29-A30. It is possible that the turn located at G29-A30 in Aβ(25–35) might play a key role in initiating the aggregation of the full-length Aβ(1–40).
We note that while the turn may play a critical role in initiating aggregation, other nucleation sites are possible. Indeed, the hydrophobic-rich C-terminus of an Aβ(25–35) peptide may readily associate through hydrophobic interactions with the C-terminus of a second peptide, leading to extended (parallel or antiparallel) dimers stabilized by H-bonds, or to dimers of hairpins (with the N-terminal regions folding over). These dimers could then grow into full-fledged fibrils. Yet another possibility is the formation of amorphous aggregates, which can act as either on- or off-pathway intermediates for fibril formation.
It is quite possible that a variety of aggregation scenarios can be realized for the Aβ(25–35) peptide. Indeed, experiments by Petkova et al. (7) demonstrate that different experimental preparation conditions led to fibrils of different morphologies. This suggests that different monomeric structures present in solution will lead to different fibril seeds (nuclei), hence resulting in different end fibrils. For our particular peptide, protofilaments of different diameters and shapes have indeed been reported, consistent with different possible nucleation scenarios. Atomic force microscope images of incubated Aβ(25–35) peptides showed that this peptide had two distinct protofilament morphologies with diameters of 1.41 ± 0.48 nm and 3.58 ± 1.53 nm, respectively (55). Taking a distance of ∼0.35 nm between adjacent residues in an extended β-conformation, the first set of diameters is compatible with the peptides adopting β-hairpin conformations over the width of the protofilaments and second set with the peptide adopting extended β-strands conformations. Structural data on Alzheimer's amyloid oligomers and fibrils, obtained from experimental (52,56–58) and computational techniques (43–45,59–61). suggest that shorter fragments assemble into β-sheets (formed of β-strands), while larger fragments (including the full-length Aβ peptides) assemble into fibrils containing hairpinlike structures. The Aβ25–35 peptide appears to be a particularly interesting case, as our simulations indicate that it could aggregate both into extended β-strand or β-hairpin aggregates.
Finally, we note that in addition to playing a role in initiating aggregation, the β-turn seen in our simulations may be responsible for the observed toxicity of soluble (monomeric or possibly small oligomeric) forms of this peptide (5,6,41). Indeed, soluble Aβ25–35 is known to bind to protein receptors on microglia, leading to their activation and subsequently to damage to neurons. β-turns are a structural motif often involved in binding to receptor proteins, and it is possible that the presence of such turns in Aβ(25–35) may be necessary to induce toxicity (41).
Acknowledgments
We thank A. Baumketner, P. Soto, and J. Higo for helpful discussions.
Support from the National Science Foundation career award No. 0133504, the A. P. Sloan Foundation, and the David and Lucile Packard foundation is gratefully acknowledged. Simulations were performed using the computational resources of the California NanoSystems Institute through National Science Foundation grant No. CHE-0321368.
References
- 1.Nunan, J., and D. H. Small. 2000. Regulation of APP cleavage by α-, β-, and γ-secretases. FEBS Lett. 483:6–10. [DOI] [PubMed] [Google Scholar]
- 2.Fraser, P. E., L. K. Duffy, M. B. O'Malley, J. Nguyen, H. Inouye, and D. A. Kirschner. 1991. Morphology and antibody recognition of synthetic β-amyloid peptides. J. Neurosci. Res. 28:474–485. [DOI] [PubMed] [Google Scholar]
- 3.Kubo, T., S. Nishimura, Y. Kumagae, and I. Kaneko. 2002. In vivo conversion of racemized β-amyloid ([d-Ser 26]A β 1–40) to truncated and toxic fragments ([d-Ser 26]A β 25–35/40) and fragment presence in the brains of Alzheimer's patients. J. Neurosci. Res. 70:474–483. [DOI] [PubMed] [Google Scholar]
- 4.Pike, C. J., A. J. Walencewicz-Wasserman, J. Kosmoski, D. H. Cribbs, C. G. Glabe, and C. W. Cotman. 1995. Structure-activity analyses of β-amyloid peptides: contributions of the β 25–35 region to aggregation and neurotoxicity. J. Neurochem. 64:253–265. [DOI] [PubMed] [Google Scholar]
- 5.Misiti, F., B. Sampaolese, M. Pezzotti, S. Marini, M. Coletta, L. Ceccarelli, B. Giardina, and M. E. Clementi. 2005. Aβ (31–35) peptide induces apoptosis in pc 12 cells: contrast with Aβ (25–35) peptide and examination of underlying mechanisms. Neurochem. Int. 46:575–583. [DOI] [PubMed] [Google Scholar]
- 6.Clementi, M. E., S. Marini, M. Coletta, F. Orsini, B. Giardina, and F. Misiti. 2005. Aβ(31–35) and Aβ(25–35) fragments of amyloid β-protein induce cellular death through apoptotic signals: role of the redox state of methionine-35. FEBS Lett. 579:2913–2918. [DOI] [PubMed] [Google Scholar]
- 7.Petkova, A. T., R. D. Leapman, Z. Guo, W. M. Yau, M. P. Mattson, and R. Tycko. 2005. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 307:262–265. [DOI] [PubMed] [Google Scholar]
- 8.Kohno, T., K. Kobayashi, T. Maeda, K. Sato, and A. Takashima. 1996. Three-dimensional structures of the amyloid β peptide (25–35) in membrane-mimicking environment. Biochemistry. 35:16094–16104. [DOI] [PubMed] [Google Scholar]
- 9.D'Ursi, A. M., M. R. Armenante, R. Guerrini, S. Salvadori, G. Sorrentino, and D. Picone. 2004. Solution structure of amyloid β-peptide (25–35) in different media. J. Med. Chem. 47:4231–4238. [DOI] [PubMed] [Google Scholar]
- 10.Terzi, E., G. Holzemann, and J. Seelig. 1994. Reversible random coil-β-sheet transition of the Alzheimer β-amyloid fragment (25–35). Biochemistry. 33:1345–1350. [DOI] [PubMed] [Google Scholar]
- 11.Terzi, E., G. Holzemann, and J. Seelig. 1994. Alzheimer β-amyloid peptide 25–35: electrostatic interactions with phospholipid membranes. Biochemistry. 33:7434–7441. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko, I., N. Yamada, Y. Sakuraba, M. Kamenosono, and S. Tutumi. 1995. Suppression of mitochondrial succinate dehydrogenase, a primary target of β-amyloid, and its derivative racemized at Ser residue. J. Neurochem. 65:2585–2593. [DOI] [PubMed] [Google Scholar]
- 13.El-Agnaf, O. M., G. B. Irvine, G. Fitzpatrick, W. K. Glass, and D. J. Guthrie. 1998. Comparative studies on peptides representing the so-called tachykinin-like region of the Alzheimer Aβ peptide (Aβ(25–35)). Biochem. J. 336:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shanmugam, G., and P. M. Polararapu. 2004. Structure of Aβ(25–35) peptide in different environment. Biophys. J. 622:622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Agnaf, O. M., G. B. Irvine, and D. J. Guthrie. 1997. Conformations of β-amyloid in solution. J. Neurochem. 68:437–439. [PubMed] [Google Scholar]
- 16.Berendsen, H. J. C. 1981. Intermolecular forces. In Interaction Models for Water in Relation to Protein Hydration. Reidel, Dordrecht, The Netherlands.
- 17.Fioroni, M., K. Burger, A. E. Mark, and D. Roccatano. 2001. Model of 1,1,1,3,3,3-hexafluoro-propan-2-ol for molecular dynamics simulations. J. Phys. Chem. B. 105:10967–10975. [Google Scholar]
- 18.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 91:43–56. [Google Scholar]
- 19.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 7:306–317. [Google Scholar]
- 20.van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hunenberger, P. Kruger, A. E. Mark, W. R. P. Scott, and I. G. Tironi. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Vdf Hochschulverland, ETH, Zurich, Switzerland.
- 21.Berendsen, H. J. C., J. P. M. Postma, A. Di Nola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 22.Okamoto, Y. 2004. Generalized-ensemble algorithms: enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J. Mol. Graph. Mod. 22:425–439. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, K., and J. Higo. 2003. Free-energy landscape of a chameleon sequence in explicit water and its inherent α/β bifacial property. Protein Sci. 12:2542–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nymeyer, H., S. Gnanakaran, and A. E. Garcia. 2004. Atomic simulations of protein folding, using the replica exchange algorithm. Methods Enzymol. 383:119. [DOI] [PubMed] [Google Scholar]
- 25.Sugita, Y., and Y. Okamoto. 1999. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314:141. [Google Scholar]
- 26.Pitera, J. W., and W. Swope. 2003. Understanding folding and design: replica-exchange simulations of “Trp-cage” miniproteins. Proc. Natl. Acad. Sci. USA. 100:7587–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia, A. E., and J. N. Onuchic. 2003. Folding a protein in a computer: an atomic description of the folding/unfolding of protein A. Proc. Natl. Acad. Sci. USA. 100:13898–13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch, W. S. C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 29.Daura, X., K. Gademann, B. Jaun, D. Seebach, A. E. van Gunsteren, and W. F. Mark. 1999. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. Engl. 38:236–240. [Google Scholar]
- 30.Jasanoff, A., and A. R. Fersht. 1994. Quantitative determination of helical propensities from trifluoroethanol titration curves. Biochemistry. 33:2129–2135. [DOI] [PubMed] [Google Scholar]
- 31.Diaz, M. D., and S. Berger. 2001. Preferential solvation of a tetrapeptide by trifluoroethanol as studied by intermolecular NOE. Magn. Reson. Chem. 39:369–373. [Google Scholar]
- 32.Fioroni, M., M. D. Diaz, K. Burger, and S. Berger. 2002. Solvation phenomena of a tetrapeptide in water/trifluoroethanol and water/ethanol mixtures: a diffusion NMR, intermolecular NOE, and molecular dynamics study. J. Am. Chem. Soc. 124:7737–7744. [DOI] [PubMed] [Google Scholar]
- 33.Gerig, J. T. 2004. Structure and solvation of melittin in 1,1,1,3,3,3-hexafluoro-2-propanol/water. Biophys. J. 86:3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vila, J. A., D. R. Ripoll, and H. A. Scheraga. 2000. Physical reasons for the unusual α-helix stabilization afforded by charged or neutral polar residues in alanine-rich peptides. Proc. Natl. Acad. Sci. USA. 97:13075–13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh, T., S. Garde, and A. E. Garcia. 2003. Role of backbone hydration and salt-bridge formation in stability of α-helix in solution. Biophys. J. 85:3187–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks, C. L. R., and L. Nilsson. 1993. Promotion of helix formation in peptides dissolved in alcohol and water-alcohol mixtures. J. Am. Chem. Soc. 115:11034–11035. [Google Scholar]
- 37.Roccatano, D., M. Fioroni, M. Zacharias, and G. Colombo. 2005. Effect of hexafluoroisopropanol alcohol on the structure of melittin: a molecular dynamics simulation study. Protein Sci. 14:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daidone, I., F. Simona, D. Roccatano, R. A. Broglia, G. Tiana, G. Colombo, and A. Di Nola. 2004. β-hairpin conformation of fibrillogenic peptides: structure and α-β transition mechanism revealed by molecular dynamics simulation. Proteins. 57:198–204. [DOI] [PubMed] [Google Scholar]
- 39.Hardy, J., and D. J. Selkoe. 2002. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 297:353–357. [DOI] [PubMed] [Google Scholar]
- 40.Temussi, P. A., L. Masino, and A. Pastore. 2003. From Alzheimer to Huntington: why is a structural understanding so difficult? EMBO J. 22:355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashioka, S., A. Monji, T. Ueda, S. Kanba, and H. Nakanishi. 2005. Amyloid-β fibril formation is not necessarily required for microglial activation by the peptides. Neurochem. Int. 47:369–376. [DOI] [PubMed] [Google Scholar]
- 42.Haque, T. S., J. Little, and S. H. Gellman. 1996. Stereochemical requirements for β-hairpin formation: model studies with four-residue peptides and depsipeptides. J. Am. Chem. Soc. 118:6975–6985. [Google Scholar]
- 43.Baumketner, A., and J.-E. Shea. 2005. Free energy landscapes for amyloidogenic tetrapeptides dimerization. Biophys. J. 89:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klimov, D., and D. Thirumalai. 2003. Dissecting the assembly of Aβ16–22 amyloid peptides into antiparallel β-sheets. Structure. 11:295–307. [DOI] [PubMed] [Google Scholar]
- 45.Gnanakaran, S., R. Nussinov, and A. E. Garcia. 2006. Atomic-level description of amyloid β dimer formation. J. Am. Chem. Soc. 128:2158–2159. [DOI] [PubMed] [Google Scholar]
- 46.Nishino, M., Y. Sugita, T. Yoda, and Y. Okamoto. 2005. Structures of a peptide fragment of β2-microglobulin studied by replica-exchange molecular dynamics simulations—towards the understanding of the mechanism of amyloid formation. FEBS Lett. 579:5425–5429. [DOI] [PubMed] [Google Scholar]
- 47.Han, W., and Y. D. Wu. 2005. A strand-loop-strand structure is a possible intermediate in fibril elongation: long time simulations of amyloid-β peptide (10–35). J. Am. Chem. Soc. 127:15408–15416. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted in proof.
- 49.Baumketner, A., S. L. Bernstein, T. Wyttenbach, G. Bitan, D. B. Teplow, and J.-E. Shea. 2006. Amyloid β-protein monomer structure: a computational and experimental study. Protein Sci. 15:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flock, D., S. Colacino, G. Colombo, and A. Di Nola. 2006. Misfolding of the amyloid-β protein: a molecular dynamics study. Protein Struct. Funct. Bioinf. 62:183–192. [DOI] [PubMed] [Google Scholar]
- 51.Fernández, A., and H. A. Scheraga. 2003. Insufficiently dehydrated hydrogen bonds as determinants of protein interactions. Proc. Natl. Acad. Sci. USA. 100:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petkova, A. T., Y. Ishii, J. J. Balbach, O. N. Antzutkin, R. D. Leapman, F. Delaglio, and R. Tycko. 2002. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 99:16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, A. D., E. Portelius, I. Kheterpal, J. Guo, K. D. Kelsey, D. Cook, Y. Xu, and R. Wetzel. 2004. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 335:833–842. [DOI] [PubMed] [Google Scholar]
- 54.Williams, A. D., S. Shivaprasad, and R. Wetzel. 2006. Alanine scanning mutagenesis of Aβ(1–40) amyloid fibril stability. J. Mol. Biol. 357:1283–1294. [DOI] [PubMed]
- 55.Liu, R., C. McAllister, Y. Lyubchenko, and M. R. Sierks. 2004. Residues 17–20 and 30–35 of β-amyloid play critical roles in aggregation. J. Neurosci. Res. 75:162–171. [DOI] [PubMed] [Google Scholar]
- 56.Balbach, J. J., Y. Ishii, O. N. Antzutkin, R. D. Leapman, N. W. Rizzo, F. Dyda, J. Reed, and R. Tycko. 2000. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 39:13748–13759. [DOI] [PubMed] [Google Scholar]
- 57.Luhrs, T., C. Ritter, M. Adrian, D. Riek-Loher, B. Bohrmann, H. Dobeli, D. Schubert, and R. Riek. 2005. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. USA. 102:17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chimon, S., and Y. Ishii. 2005. Capturing intermediate structures of Alzheimer's β-amyloid, Aβ(1–40), by solid-state NMR spectroscopy. J. Am. Chem. Soc. 127:13472–13473. [DOI] [PubMed] [Google Scholar]
- 59.Ma, B., and R. Nussinov. 2002. Stabilities and conformations of Alzheimer's β-amyloid peptide oligomers (Aβ16–22,Aβ10–35): sequence effects. Proc. Natl. Acad. Sci. USA. 99:14126–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Favrin, G., A. Irback, and S. Mohanty. 2004. Oligomerization of amyloid Aβ16–22 peptides using hydrogen bonds and hydrophobicity forces. Biophys. J. 87:3657–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santini, S., G. Wei, N. Mousseau, and P. Derreumaux. 2004. Pathway complexity of Alzheimer's β-amyloid Aβ16–22 peptide-peptide assembly. Structure. 12:1245–1255. [DOI] [PubMed] [Google Scholar]