Abstract
Transcription in eukaryotes is influenced by the chromatin state of the template, and chromatin remodeling factors have well-documented roles in regulating transcription initiation by RNA polymerase (pol) II. Chromatin also influences transcription elongation; however, little is known about the role of chromatin remodeling factors in this process. Here, we present evidence that the Saccharomyces cerevisiae chromatin remodeling factor Chd1 functions during transcription elongation. First, we identified Chd1 in a two-hybrid screen for proteins that interact with Rtf1, a member of the Paf1 complex that associates with RNA pol II and regulates transcription elongation. Secondly, we show through co-immunoprecipitation studies that Chd1 also interacts with components of two essential elongation factors, Spt4–Spt5 and Spt16–Pob3. Thirdly, we demonstrate that deletion of CHD1 suppresses a cold-sensitive spt5 mutation that is also suppressed by defects in the Paf1 complex and RNA pol II. Finally, we demonstrate that Chd1, Rtf1 and Spt5 associate with actively transcribed regions of chromatin. Collectively, these findings suggest an important role for Chd1 and chromatin remodeling in the control of transcription elongation.
Keywords: Chd1/chromatin remodeling/Paf1 complex/Spt4–Spt5/transcription elongation
Introduction
The packaging of DNA into chromatin presents a significant impediment to transcription initiation and elongation by RNA polymerase (pol) II. Transcription initiation is often regulated by proteins that manipulate chromatin structure and alter the accessibility of promoter DNA. Chromatin structure over promoters typically is regulated either by proteins that alter chromatin by ATP-driven mechanisms or by post-translational modification of histones (Narlikar et al., 2002). Similarly, RNA pol II is thought to use accessory factors to facilitate its progression through nucleosomes during the elongation phase of transcription (Hartzog et al., 2002). Several RNA pol II-associated protein complexes have been proposed to regulate transcription elongation through an effect on chromatin; however, the mechanisms by which these factors function remain obscure.
The activities of two essential transcription elongation factors in Saccharomyces cerevisiae, Spt4–Spt5 and Spt16/Cdc68–Pob3 (hereafter, referred to as Spt16– Pob3), appear to be linked to chromatin. Compo nents of these complexes were identified originally by genetic screens for transcriptional regulators in yeast, and related transcription factor complexes have been characterized in mammalian cells where Spt4–Spt5 is known as DSIF and Spt16–Pob3 is known as FACT (Hartzog et al., 2002). Biochemical experiments with yeast or human proteins have revealed interactions of RNA pol II with Spt4–Spt5 and Spt16–Pob3 and demonstrated negative as well as positive effects on transcription elongation (Bourgeois et al., 2002; Hartzog et al., 2002; Krogan et al., 2002; Lindstrom et al., 2003). These observations are supported by genetic data that indicate functional connections between RNA pol II, Spt4–Spt5, Spt16–Pob3 and other proteins involved in the control of transcription elongation (Hartzog et al., 1998; Orphanides et al., 1999; Costa and Arndt, 2000; Lindstrom and Hartzog, 2001; Squazzo et al., 2002). Moreover, Spt5 co-localizes with the hyperphosphorylated, elongation-competent form of RNA pol II on Drosophila polytene chromosomes (Kaplan et al., 2000), and chromatin immunoprecipitation (ChIP) studies place Spt5 on actively transcribed genes in Drosophila and yeast (Andrulis et al., 2000; Pokholok et al., 2002). The first indications that the functions of Spt4–Spt5 and Spt16–Pob3 were connected to chromatin came from phenotypic similarities caused by mutations in SPT4, SPT5, SPT16 and genes that encode histones (Winston, 1992; Hartzog et al., 2002). Later, Spt16– Pob3 (in association with Nhp6) and FACT were found to interact genetically and physically with histones (Orphanides et al., 1999; Formosa et al., 2002), and FACT was shown to stimulate transcription elongation on a chromatin-assembled template in vitro (Orphanides et al., 1998, 1999).
Another factor implicated in transcription elongation is the Paf1 complex (Mueller and Jaehning, 2002; Pokholok et al., 2002; Squazzo et al., 2002). Minimally comprised of Paf1, Ctr9, Cdc73, Rtf1 and Leo1 (Mueller and Jaehning, 2002; Squazzo et al., 2002), this complex was identified originally as an RNA pol II-interacting complex distinct from the Srb mediator (Shi et al., 1997). Recent genetic and biochemical results strongly suggest a role for the Paf1 complex in transcription elongation. These findings include the association of the Paf1 complex with Spt4–Spt5 and Spt16–Pob3 (Gavin et al., 2002; Krogan et al., 2002; Mueller and Jaehning, 2002; Squazzo et al., 2002; Lindstrom et al., 2003), genetic suppression of spt5 mutations by mutations in PAF1 and LEO1 (Squazzo et al., 2002), and strong genetic interactions between members of the Paf1 complex and other factors involved in transcription elongation (Costa and Arndt, 2000; Formosa et al., 2002; Squazzo et al., 2002). In certain genetic backgrounds, deletion of members of the Paf1 complex causes sensitivity to 6-azauracil (6AU) and mycophenolic acid, two phenotypes frequently associated with defects in transcription elongation (Costa and Arndt, 2000; Desmoucelles et al., 2002; Krogan et al., 2002; Squazzo et al., 2002). In addition, members of the Paf1 complex associate with the coding regions of genes in vivo (Krogan et al., 2002; Pokholok et al., 2002). These data indicate the existence of one or more RNA pol II elongation complexes that include Spt4–Spt5, Spt16– Pob3 and the Paf1 complex and have the potential to mediate interactions of RNA pol II with chromatin during transcription elongation.
In this work, we describe evidence for an additional protein that associates with these elongation factors, Chd1. CHD proteins comprise a highly conserved, yet poorly understood, class of chromatin remodeling factor with an intriguing tripartite domain structure (Woodage et al., 1997). Chd1 and other CHD proteins have two chromodomains near the N-terminus, a centrally located Snf2-related helicase/ATPase domain and a Myb-related DNA-binding domain near the C-terminus (Woodage et al., 1997; Figure 1A). Chromodomains can play important roles in localizing proteins to chromatin (Eissenberg, 2001), and the Chd1 proteins of mouse and Drosophila are chromatin associated (Stokes and Perry, 1995; Stokes et al., 1996; Kelley et al., 1999). Consistent with the presence of an Snf2-related helicase/ATPase domain, S.cerevisiae Chd1 can remodel nucleosome structure in vitro in an ATP-dependent manner (Tran et al., 2000). However, despite the presence of three conserved domains implicated in chromatin binding and remodeling, chd1 mutations cause only weak mutant phenotypes in yeast (Woodage et al., 1997). Hence, the in vivo function of Chd1 is unknown.
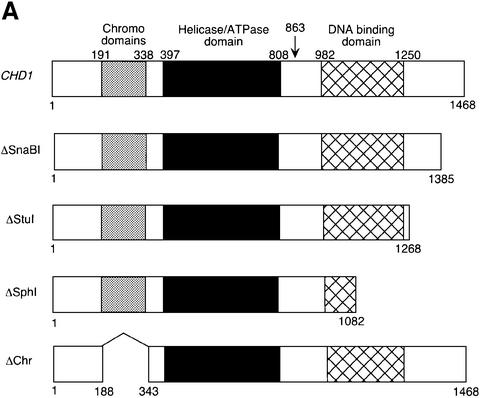
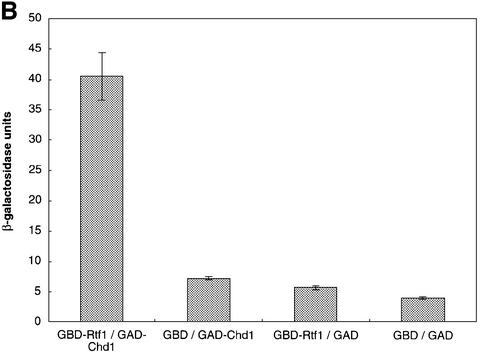
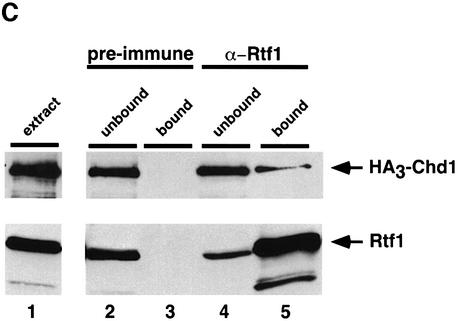
Fig. 1. Identification of Chd1 as an Rtf1-interacting protein. (A) Diagram of the wild-type Chd1 protein and deletion derivatives used in the analysis shown in Table I and Figure 2. The plasmids encoding these different forms of Chd1 are: CHD1, pGH269; ΔSnaBI, pGH272; ΔStuI, pGH273; ΔSphI, pGH274; and ΔChr, pGH271. The Gal4–Chd1 fusion protein identified in the two-hybrid screen with Rtf1 contains amino acids 863 (arrow) to 1468. (B) β-galactosidase assays were performed on transformants of yeast strain PJ69-4A that expressed the indicated Gal4 fusion proteins (GBD, Gal4-DNA binding domain; GAD, Gal4 activation domain). The following plasmids were used: pLS28 (GBD-Rtf1), pKA202 (original library isolate of GAD-Chd1), pGBT9 and pGAD424. For each plasmid combination, the reported units are mean values from assays performed on three independent transformants at two different extract concentrations. Error bars denote standard errors. (C) Chd1 co-immunoprecipitates with Rtf1. Anti-Rtf1 or pre-immune serum was added to extracts prepared from yeast strain GHY773 for co-immunoprecipitation analysis. Precipitated proteins were analyzed by immunoblotting with anti-Rtf1 and anti-HA1 antisera. Lane 1, 20 µg of whole-cell extract; lanes 2 and 4, 20 µg of unbound material; lanes 3 and 5, total bound material.
Here, we report a number of observations suggesting a role for Chd1 in transcription elongation by RNA pol II. First, we identify a two-hybrid interaction between Rtf1 and Chd1. Secondly, we show that Chd1 co-immunoprecipitates with members of the Paf1, Spt4–Spt5 and Spt16–Pob3 complexes. Thirdly, we show that a chd1 deletion mutation suppresses an spt5 cold-sensitive mutation, and we further use genetics to show that all three of the conserved domains of Chd1 are required for its function in vivo. Finally, we show that Rtf1, Spt5 and Chd1 can be localized to chromatin that is being transcribed by RNA pol II. These findings provide strong evidence that Chd1 functions in the elongation phase of transcription and provide a platform for future analysis of the role of ATP-dependent chromatin remodeling factors and chromodomain proteins in elongation.
Results
Chd1 physically associates with Rtf1
By affinity purification and co-immunoprecipitation analysis, we previously demonstrated that Rtf1 is a member of the RNA pol II-associated Paf1 complex (Squazzo et al., 2002). As a complementary approach toward identifying proteins that interact with Rtf1, we performed a two-hybrid screen with a plasmid encoding full-length Rtf1 fused to the DNA-binding domain of Gal4. The screen employed a library of Gal4 activation domain fusion plasmids and a yeast strain that harbors three different Gal4-responsive reporter genes (James et al., 1996). The plasmid encoding the strongest candidate for an Rtf1-interacting protein, as judged by expression of all three reporter genes, was analyzed by sequencing and was found to encode a Gal4–Chd1 fusion protein that includes amino acids 863–1468 of Chd1. This C-terminal region of Chd1 contains the Myb-related DNA-binding domain but lacks the chromo- and helicase domains (Figure 1A). β-galactosidase assays on liquid-grown cultures confirmed that the two-hybrid interaction was dependent on both the Rtf1 and Chd1 sequences (Figure 1B).
To test the interaction between Rtf1 and Chd1 by an independent method, we immunoprecipitated Rtf1 from extracts of yeast cells that expressed an HA3 epitope-tagged form of Chd1 and analyzed the co-precipitates by immunoblotting. As shown in Figure 1C, a fraction of the cellular pool of HA3-Chd1 reproducibly co-precipitated with Rtf1 from these extracts. The precipitation of HA3-Chd1 was not detected when pre-immune serum was used (Figure 1C) or when extracts were prepared from an rtf1Δ strain (data not shown). Together with the results of the two-hybrid screen, these results indicate a physical association between Chd1 and Rtf1 in vivo.
Chd1 interacts genetically and physically with the transcription elongation factor Spt5
Previously, we found that a specific spt5 mutation, spt5-242, was genetically suppressed by mutations in PAF1, LEO1 and genes that encode RNA pol II subunits (Hartzog et al., 1998; Squazzo et al., 2002). Consistent with these genetic findings, Spt5 associates with both RNA pol II and the Paf1 complex (Hartzog et al., 1998; Wada et al., 1998; Mueller and Jaehning, 2002; Squazzo et al., 2002). Although chd1 mutations were not identified in our original genetic selection, we have found that chd1 deletion mutations can also suppress the spt5-242 mutation (see Materials and methods; Figure 2A). Frequently, genetic interactions occur between genes that encode interacting proteins. We therefore prepared extracts from strains expressing a Flag epitope-tagged form of Spt5 and HA3-Chd1 and performed anti-Flag and anti-HA immunoprecipitations. In both experiments, we found that Spt5 and Chd1 specifically co-precipitate from yeast cell extracts and that this co-precipitation depended upon the presence of the appropriate epitope on the target protein (Figure 2B and C). Furthermore, consistent with a previously reported genetic interaction between POB3 and CHD1 (Costa and Arndt, 2000) and recent affinity purification results (Krogan et al., 2002), we observed that Pob3 specifically co-immunoprecipitated with HA3-Chd1 (Figure 2C). In contrast, the TATA-binding protein (TBP), which functions only in initiation, did not co-precipitate with HA3-Chd1 (data not shown). These results support the identification of Chd1 as an Rtf1-interacting protein and further strengthen the functional and physical connections between the Paf1 complex and the Spt4–Spt5 and Spt16–Pob3 transcription elongation factors.
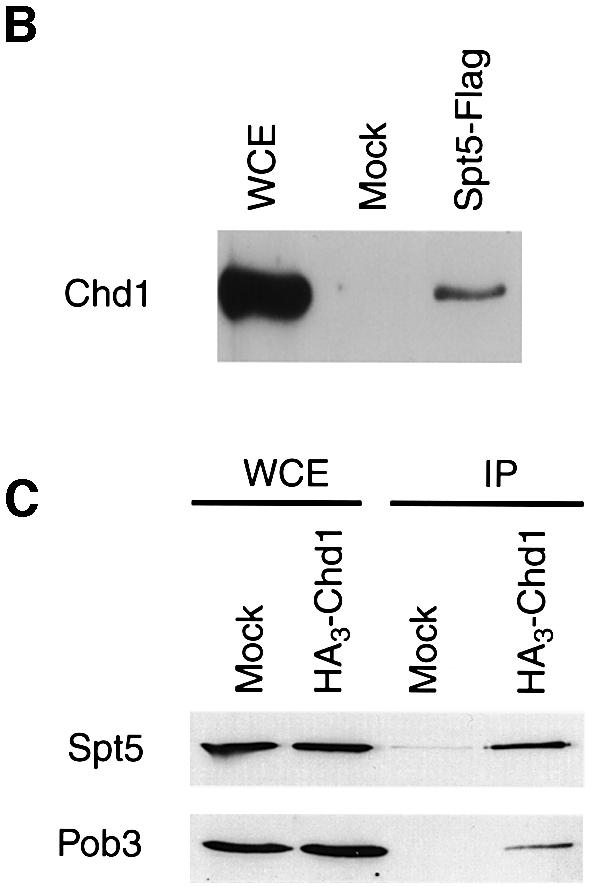
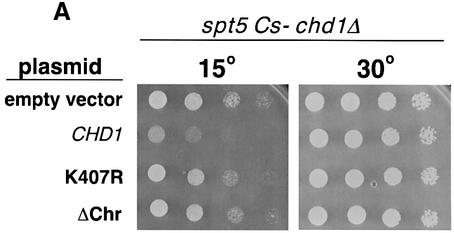
Fig. 2. Spt5 and Chd1 interactions. (A) chd1Δ mutations suppress an spt5 Cs– mutation. A chd1Δ spt5 Cs– strain (GHY305) was transformed with plasmids encoding wild-type Chd1 (pGH269), Chd1 with the K407R substitution (pGH268), a derivative of Chd1 lacking both chromodomains (pGH271), and an empty URA3 vector (pRS316). Five-fold dilutions of the transformants were plated to two SC-uracil plates. One of the plates was incubated at 15°C for 6 days and the other was incubated at 30°C for 2 days. (B) Chd1 co-immunoprecipitates with Spt5-Flag. Anti-Flag immunoprecipitations were performed on extracts of an Spt5-Flag strain (GHY617) and an untagged control strain (mock, GHY611). Precipitated proteins were analyzed by immunoblotting with anti-Chd1 antisera (Tran et al., 2000). Lanes: WCE, 30 µg of the Spt5-Flag extract; Mock, the untagged immunoprecipitate; Spt5-Flag, the Spt5-Flag immunoprecipitate. (C) Spt5 and Pob3 co-immunoprecipitate with HA3-Chd1. Extracts of a HA3-Chd1strain (GHY773) and an untagged control strain (mock; FY119) were immunoprecipitated with anti-HA antisera. Precipitated proteins were analyzed by immunoblotting with anti-Spt5 and anti-Pob3 antisera.
Genetic analysis of the conserved domains of CHD1
Few phenotypes have been attributed to mutations in CHD1, limiting an understanding of Chd1 function in vivo. Consequently, we exploited the spt5 suppression phenotype to analyze chd1 mutations and identify regions of the gene important for its function. A set of Chd1 deletion mutants was generated that lacked regions from the C-terminus or the two chromodomains of Chd1 (Figure 1A). In addition, we introduced a single amino acid substitution, K407R, into Chd1. Lys407 falls within an adenine nucleotide-binding motif, GXGKT, that is shared by many proteins requiring ATP for their function (Walker et al., 1982). Similar substitutions in other chromatin remodeling proteins result in the abolition of their ATPase and remodeling activities as well as their in vivo functions (see Corona et al., 1999; and references therein). Immunoblot analyses revealed that each of these mutant proteins was expressed to near wild-type levels and migrated with the expected mobility on SDS–gels (data not shown). Each of the chd1 mutations was tested for its ability to complement a chd1Δ mutation in a strain containing the Cs– spt5-242 allele (Figure 2A; Table I). The spt5-242 chd1Δ strain is able to grow at 15°C, and the mutants were assayed for their ability to revert the strain to a Cs– phenotype. Although this phenotype was complemented by the plasmid-borne HA3-CHD1 gene, none of the C-terminal truncation mutants was able to complement the chd1Δ mutation for spt5 suppression (Figure 2A; Table I). Moreover, deletion of the chromodomains or introduction of the K407R change also prevented complementation (Figure 2A; Table I). A second phenotype associated with loss of CHD1 is a Ts– growth defect at 37°C observed in isw1Δ isw2Δ chd1Δ triple mutants (Table I and Tsukiyama et al., 1999). This Ts– phenotype was complemented by the plasmid-borne HA3-CHD1, and partially complemented by the ΔSnaBI deletion allele of CHD1, but not by any other chd1 deletion or point mutation (Table I). Taken together, these results suggest that all three conserved motifs within Chd1 are necessary for its activity in vivo. These findings are consistent with previous mutational studies performed on mouse Chd1 that investigated the importance of these domains for nuclear localization (Kelley et al., 1999).
Table I. All three conserved domains of Chd1 are required for its function.
| Plasmida | spt5-242 chd1Δb | isw1Δ isw2Δ chd1Δc |
|---|---|---|
| growth at 15°C | growth at 37°C | |
| Vector | +/– | – |
| CHD1 | – | + |
| ΔSnaBI | +/– | +/– |
| ΔStuI | +/– | – |
| ΔSphI | +/– | – |
| ΔChr | +/– | – |
| K407R | +/– | – |
aMutant Chd1 proteins encoded by the indicated plasmids are diagrammed in Figure 1.
bStrain GHY305.
cStrain YTT227.
+ indicates growth of clearly visible large colonies; +/– indicates poorly growing small colonies; –indicates no visible individual colonies observed. Growth at 37°C was scored after 2 days, and growth at 15°C was scored after 6 days.
Rtf1 and Spt5 associate with the coding regions of actively transcribed genes
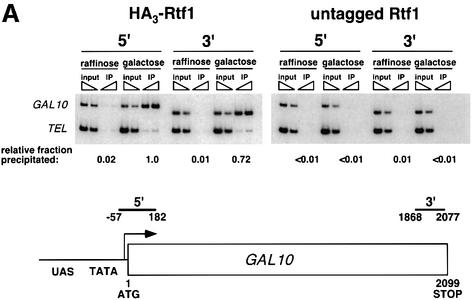
Previous studies provided strong functional and biochemical links between the Paf1 complex, the Spt4–Spt5 complex and RNA pol II. To investigate these connections further and to ascertain the locations of Rtf1 and Spt5 on actively transcribed genes, we performed ChIP studies with functional, epitope-tagged forms of the proteins. Using an HA3-tagged derivative of Rtf1 and a Flag-tagged derivative of Spt5, we investigated the location of Rtf1 and Spt5 on several RNA pol II-transcribed genes (Figure 3). On the galactose-inducible GAL10 gene, HA3-Rtf1 cross-linked to sequences at both the 5′ and 3′ ends of the open reading frame (ORF) in a manner strongly dependent upon active transcription and upon the HA3 tag (Figure 3A). In the absence of galactose (raffinose-grown cells) when GAL10 is not expressed, HA3-Rtf1 could not be detected at either end of the gene. Consistent with recently published observations, we also observed a galactose-dependent recruitment of Spt5 to the ORF of the GAL1 gene (Pokholok et al., 2002; data not shown).
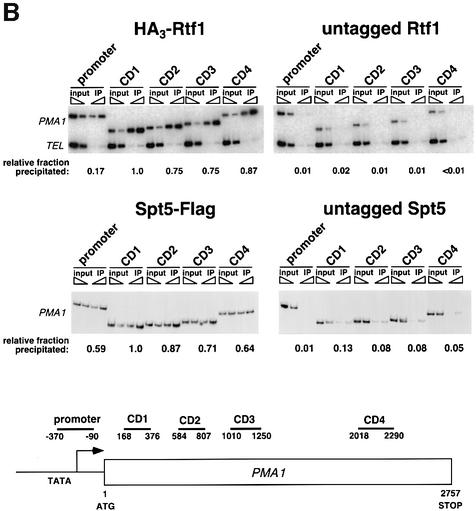
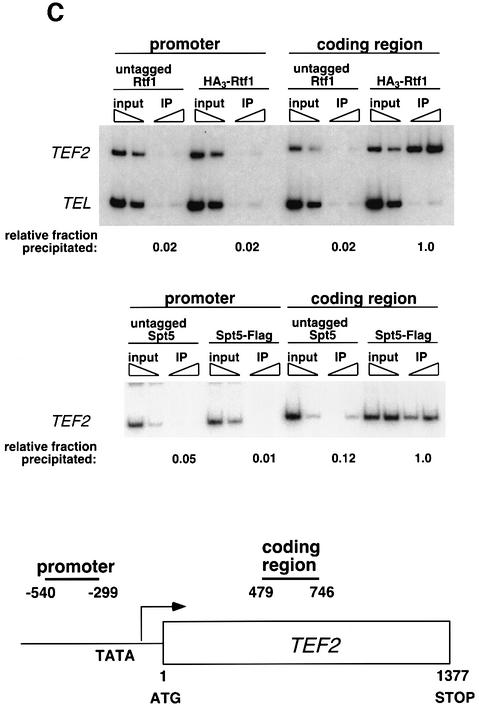
Fig. 3. Rtf1 and Spt5 associate with the coding regions of genes. (A) ChIP analysis of Rtf1 on the GAL10 gene was performed on transformants of yeast strain KY399 that expressed HA3-Rtf1 or untagged Rtf1. Strains were grown in media containing raffinose, and GAL gene transcription was induced by addition of 2% galactose for 20 min. Cross-linked chromatin was immunoprecipitated with anti-HA1 antibody, and PCR was conducted on two dilutions of input DNA and two different amounts of precipitated DNA, as indicated. Locations of the 5′ and 3′ primer pairs, relative to the ATG, are given at the bottom. (B) ChIP analysis of Rtf1 and Spt5 on the PMA1 gene. For Rtf1, chromatin isolated from glucose-grown cultures was subjected to ChIP with antibody against HA1 as described in (A). For Spt5, chromatin was isolated from glucose-grown cultures of strains GHY1300 (Spt5-Flag) and FY118 (Spt5). Promoter and coding region primer sequences were described previously (Cho et al., 2001), and the locations of the predicted PCR products are shown at the bottom. (C) ChIP analysis of Rtf1 and Spt5 on the TEF2 gene. Chromatin isolated from glucose-grown cultures was subjected to ChIP as in (B). Locations of promoter (TEF2 set 2) and coding region (TEF2 set 5) primer sequences, relative to the ATG, are given below. For each ChIP experiment, PCR products were quantitated by phosphoimager analysis, divided by the corresponding input DNA signal, and normalized arbitrarily to the sample with the highest percentage input value. Subtelomeric primer sequences from chromosome VI (Vogelauer et al., 2000) were used as a control.
To determine whether the chromatin association of Rtf1 and Spt5 is limited to the GAL genes, we extended the ChIP analysis to the PMA1 and TEF2 genes. Consistent with our ChIP results on the GAL genes, HA3-Rtf1 and Flag-Spt5 associated with the PMA1 gene throughout the ORF (Figure 3B). We did not observe a large reduction in signal at the 3′ end of the coding sequence (CD4) relative to the 5′ end of the coding sequence (CD1). Within the limits of ChIP analysis, this observation suggests that Rtf1 and Spt5 may accompany RNA pol II along the entire transcription unit. We also analyzed the location of TBP on GAL10 and PMA1 to control for the sonication conditions in our ChIP procedure. We observed specific enrichment of TBP only at the 5′ ends/promoter regions of these genes (data not shown). In accordance with our findings at the GAL genes and PMA1, we also observed strong association of HA3-Rtf1 and Spt5-Flag with the coding sequence of TEF2 (Figure 3C). Together with the results of previous ChIP analyses (Andrulis et al., 2000; Krogan et al., 2002; Pokholok et al., 2002), these results strongly suggest that the Paf1 complex and Spt5 associate with actively transcribing RNA pol II at multiple genes in vivo.
Chd1 localizes to the coding regions of genes
The biochemical and genetic interactions between Chd1, Rtf1 and Spt5 suggested that Chd1 might play a role in transcription elongation and consequently associate with the transcribed regions of genes. In an attempt to uncover the genomic localization of Chd1 and to identify genes that are directly regulated by this remodeling factor, ChIP followed by microarray analysis was carried out on strains expressing a Myc6-Chd1 fusion. Our preliminary analysis of two independent experiments revealed preferential precipitation of DNA sequences from 16 loci by Myc6-Chd1 (by a factor of ≥2.5-fold relative to a reference sample; data not shown). Of the 16 loci, five corresponded to intergenic regions and 11 to the coding regions of genes. A significant number of the coding region sequences (six out of 11) were from highly expressed genes with a role in protein synthesis, including the RPL genes and TEF2.
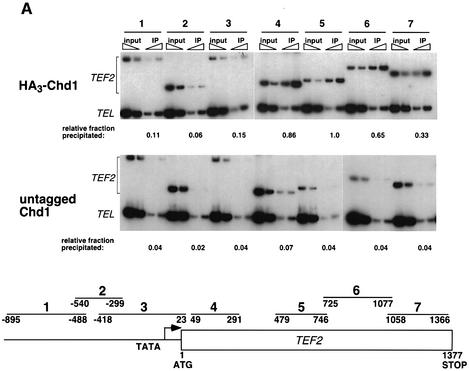
To verify the recruitment of Chd1 to a gene detected by our genome-wide localization studies, we performed quantitative ChIP analysis on TEF2 using an HA3-tagged form of Chd1 (Figure 4A). The PCR analysis showed that Chd1 preferentially cross-linked to the entire coding region of TEF2, although we consistently observed decreased precipitation of sequences near the 3′ end of the gene. Chd1 did not cross-link efficiently to sequences upstream of the ORF and, in contrast to the behavior of known regulators of TEF2 and RP genes, Chd1 was not enriched at the Rap1-binding sites (Reid et al., 2000). We also investigated the presence of Chd1 at an additional eight RP genes, some of which were not identified by the microarray experiment, and observed preferential precipitation of the coding regions of these genes by Chd1 (data not shown).
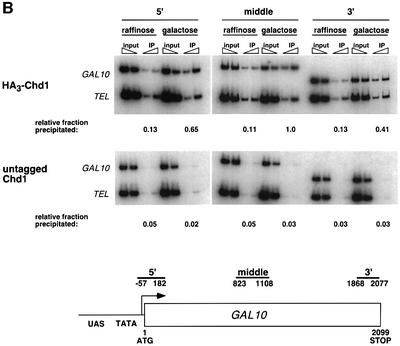
Fig. 4. Chd1 associates with the coding regions of transcribed genes. (A) ChIP analysis of HA3-Chd1 on the TEF2 gene. Strains GHY773 and FY118 were grown in YPD media and subjected to ChIP analysis with anti-HA1 antibody. Seven primer pairs that amplify the TEF2 sequences shown at the bottom were used in quantitative PCR analysis of immunoprecipitated and total (input) chromatin samples. (B) ChIP analysis of HA3-Chd1 on the GAL10 gene. Strains GHY773 and KY661 were grown in YP-raffinose media and induced with galactose as described in Figure 3A. For TEL DNA, the relative fraction precipitated did not change with carbon source.
To characterize the functional significance of Chd1 recruitment, we examined the dependence of Chd1 localization on transcriptional activity. For these experiments, we performed ChIP analysis of HA3-Chd1 on the GAL10 gene (Figure 4B). In raffinose-grown cells, HA3-Chd1 cross-linked weakly to the GAL10 coding region and to the subtelomeric region of chromosome VI (TEL). However, upon gene induction, we observed a specific and significant enrichment of GAL10 coding sequence DNA in the immunoprecipitated samples. Similarly, we also found that the association of Chd1 with the TEF2 gene is dependent upon transcriptional activation by Rap1 (data not shown). These results suggest that Chd1 can associate with transcriptionally inactive regions of chromatin and is recruited further during the process of ongoing transcription.
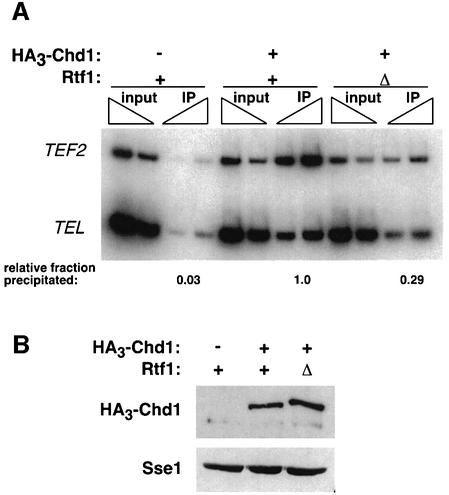
To begin to dissect the interrelationships between the various proteins that interact with elongating RNA pol II, we investigated whether the localization of Chd1 to the transcribed regions of genes was dependent on the presence of Rtf1. Therefore, we analyzed the association of HA3-Chd1 with the coding region of TEF2 in both RTF1+ and rtf1Δ strains (Figure 5A). Consistent with the physical interaction we observed between Chd1 and Rtf1, ∼3-fold less TEF2 DNA immunoprecipitated with HA3-Chd1 in chromatin prepared from strains lacking Rtf1. This apparent decrease in the association of Chd1 with the coding region of TEF2 could not be accounted for by a reduction in Chd1 levels in rtf1Δ strains (Figure 5B), nor by reduced TEF2 transcription (K.M.Arndt and K.Kumer, unpublished data). These results suggest that the interaction between Chd1 and transcribed chromatin is partially dependent on Rtf1 and may be regulated by RNA pol II-associated elongation factors.
Fig. 5. Chd1 association with TEF2 is partially dependent upon Rtf1. (A) ChIP analysis of HA3-Chd1 on the TEF2 gene in strains lacking (Δ) or containing (+) a functional RTF1 gene. Strains GHY773, KY620 and FY118 were grown in YPD media and analyzed by ChIP using anti-HA1 antibody and the TEF2 internal primer pair 5 (see Figure 4A). FY118 contains an untagged CHD1 gene (–). Sample preparation, PCR analysis and quantitation were performed as described in Figure 3. (B) Immunoblot analysis of HA3-Chd1 levels in yeast extracts (100 µg) prepared from the same strains used in (A). The filter in the upper panel was stripped and probed with anti-Sse1 antibody to serve as a loading control.
Discussion
Members of the CHD family of proteins are remarkable for their high degree of conservation and unique structural features. The presence of chromodomains, a helicase/ATPase domain and a DNA-binding domain within the same protein suggests a role for these proteins in chromatin structure or function. In support of this prediction, previous studies demonstrated that the yeast Chd1 protein can alter the structure of nucleosomes in vitro and that a complete deletion of CHD1 can affect transcription in vivo (Tran et al., 2000). However, a molecular description of the function of Chd1 has remained elusive. In this study, we provide strong evidence for a role for Chd1 in transcription elongation.
Chd1 interacts with transcription elongation factors
Through two approaches, we identified physical interactions between Chd1 and transcription elongation factors. First, using a two-hybrid screen, we identified Chd1 as a protein that interacts with Rtf1, a member of the RNA pol II-associated Paf1 complex. Secondly, using co-immunoprecipitation, we confirmed that Chd1 interacts with Rtf1, and found that it also associates with two other transcription elongation factors, Spt5 and Pob3. Consistent with these biochemical interactions, we found that deletion of CHD1 suppresses the Cs– phenotype of a specific spt5 mutation (Figure 2) and the severe growth defect caused by a pob3 point mutation (Costa and Arndt, 2000). The finding that a chd1Δ mutation suppresses an spt5 mutation that is also suppressed by mutations in RPB1, RPB2, PAF1 and LEO1 (Hartzog et al., 1998; Squazzo et al., 2002) strongly indicates a functional role for Chd1 in transcription elongation.
Our finding of physical interactions between Chd1 and transcription elongation factors is in good agreement with observations from other laboratories. A two-hybrid screen involving an N-terminal region of mouse Chd1 detected an interaction with SSRP1, the mammalian homolog of Pob3 (Kelley et al., 1999). In addition, a proteomics study of transcription elongation factors in yeast identified a protein complex that contained Spt16–Pob3 and Chd1, although the conditions used in this study failed to uncover an interaction between Chd1, the Paf1 complex and Spt4–Spt5 (Krogan et al., 2002). Based on all of these results, we postulate the existence of a transcription elongation apparatus that associates with RNA pol II, potentially in a dynamic fashion, and contains multiple activities for altering chromatin structure. These factors are likely to function in interrelated and complex ways, as each has been implicated in both positive and negative regulation of transcription.
Genetic analysis of Chd1 function
In the absence of other mutations, deletion of CHD1 causes no strong or obvious effect on the growth properties of yeast. In an extensive survey of possible mutant phenotypes, a chd1Δ mutation was found to confer resistance to very high concentrations of 6AU, but no other mutant phenotypes (Woodage et al., 1997). The most likely explanation for these observations is a functional redundancy among chromatin remodeling activities in yeast. Indeed, two groups have reported synthetic growth defects between chd1Δ mutations and mutations that eliminate ATP-dependent chromatin remodeling factors. A chd1Δ mutation causes temperature sensitivity for growth when combined with mutations in the imitation switch genes ISW1 and ISW2, and synthetic lethality with defects in the Swi–Snf complex (Tsukiyama et al., 1999; Tran et al., 2000). The connections between Chd1 and transcription elongation reported here suggest that these chromatin remodeling factors might play a role in this stage of the transcription cycle. In accordance with this idea, a biochemical fraction containing Swi–Snf can facilitate transcription through a nucleosome-imposed elongation pause in vitro (Brown et al., 1996).
We exploited the genetic interactions of CHD1 with the spt5 Cs– mutations and isw1 isw2 deletions to perform a mutational analysis of CHD1. Mutations that deleted the chromodomains, introduced a specific amino acid change in the active site of the helicase/ATPase domain or truncated the C-terminus, including the DNA-binding domain, eliminated function. Based on the involvement of Spt5 in transcription elongation, we propose that the chromo- and helicase domains of Chd1, as well as a C-terminal domain, are required for its role in this process. Our mutational data do not address whether the DNA-binding activity imparted by the Myb-related DNA-binding domain is required, since this region of the protein also harbors sequences needed for the Rtf1 interaction, and C-terminal deletions that do not obviously disrupt the DNA-binding domain also disrupt Chd1 function (Figure 1A). Likewise, we have not assayed the importance of sequences N-terminal to the chromodomains. Given that this region of mouse Chd1 interacts with SSRP1 in a two-hybrid analysis, the analogous region of yeast Chd1 may mediate the interaction with Pob3 (Kelley et al., 1999). The requirement for the chromodomains is noteworthy because these domains have been shown to play an important role in targeting proteins to chromatin through different mechanisms (Eissenberg, 2001; Bouazoune et al., 2002). The work reported here provides a basis for the functional analysis of the chromodomain of Chd1, one of only three known chromodomain proteins in S.cerevisiae (Woodage et al., 1997; Clarke et al., 1999; Bertram and Pereira-Smith, 2001).
Chd1, Rtf1 and Spt5 localize to the transcribed regions of genes
The physical and genetic interactions between Chd1 and three different protein complexes implicated in transcription elongation suggested that Chd1 might associate with transcribed chromatin. Beginning with an unbiased genome-wide survey for Chd1 interaction sites, we found an enrichment of Chd1 on the ORFs of several highly expressed genes. By using ChIP to analyze specific genes, we discovered that Chd1 localizes to the ORFs of GAL10, TEF2 and a large number of RP genes. The transcription dependence of this localization at GAL10 and TEF2 strongly suggests that Chd1 preferentially associates with genes undergoing active transcription. These findings are consistent with the previously reported association of Drosophila Chd1 with interbands and puffed regions of polytene chromosomes (Stokes et al., 1996). Therefore, unlike two other well-studied chromodomain proteins, HP1 and Polycomb (Eissenberg, 2001), which are associated with heterochromatin and transcriptional repression, Chd1 is recruited to regions of active transcription.
Based on our combined genetic and biochemical data, we previously proposed that the Paf1 complex, together with Spt4–Spt5 and Spt16–Pob3, regulates transcription elongation. Consistent with this hypothesis, we have demonstrated the presence of Rtf1 and Spt5 on the coding regions of the GAL genes, PMA1 and TEF2. Association of these proteins with the GAL genes clearly correlates with transcriptional activity. These results are in excellent agreement with recently published ChIP assays on Spt5 and members of the Paf1 complex (Krogan et al., 2002; Pokholok et al., 2002). By analyzing the longer PMA1 gene, we have attempted to learn the distribution of Rtf1 and Spt5 along a transcription unit. Within the limits of ChIP analysis, the levels of Rtf1 and Spt5 association do not appear to change dramatically along this gene. Whether these proteins and Chd1 only interact after RNA pol II has begun elongation or whether they are recruited at the promoter is not clear.
In agreement with our data, Alén et al. (2002) have reported recently that Chd1 is localized to the coding and 3′-untranslated regions of genes. Furthermore, they report a requirement for Chd1 in transcription termination by RNA pol II and find that Chd1 may operate redundantly with the Isw1 and Isw2 chromatin remodeling factors. It is not surprising that a single factor might influence both elongation and termination by RNA pol II, since sequence elements that cause elongating RNA pol II to pause are thought to play an important role in triggering termination (Proudfoot et al., 2002).
Does Chd1 have a global or a restricted role in transcription elongation? Microarray expression studies on strains deleted for CHD1 revealed a transcriptional effect of ≥2-fold on only 2–4% of the genes in yeast (Tran et al., 2000), and our preliminary genome-wide localization studies suggested that Chd1 may not be present on all genes. However, the preferential enrichment of Chd1 on highly expressed genes may also indicate that the sensitivity of our genome-wide assay precluded detection of Chd1 on other genes. Moreover, in expression studies, a more general role for Chd1 might be obscured by redundancy with other chromatin remodeling factors. Indeed, transcriptional effects of the chd1Δ mutation on TEF2 or the RP genes were not detected in the microarray studies (Tran et al., 2000), although ChIP assays clearly demonstrate that Chd1 can interact with these genes. This observation is consistent with Chd1’s interactions with Spt5 and the Paf1 complex, which have been detected on the transcribed regions of all genes tested to date. We therefore propose that Chd1 plays a role in transcription elongation more significant than that predicted by expression studies, most likely by remodeling chromatin during transcription elongation.
Materials and methods
Yeast strains, media and genetic methods
Yeast strains were constructed by standard genetic methods of tetrad analysis or transformation (Rose et al., 1990). All FY, GHY and KY strains (Table II) are isogenic to S288C and are GAL2+. Strain YTT227 (Tsukiyama et al., 1999) is a derivative of W303. Yeast media were prepared as described previously (Rose et al., 1990).
Table II. Saccharomyces cerevisiae strains.
| Strain | Genotype |
|---|---|
| FY118 | MATa his4-912δ lys2-128δ leu2Δ1 trp1Δ63 ura3-52 |
| FY119 | MATα his4-912δ lys2-128δ leu2Δ1 trp1Δ63 ura3-52 |
| GHY305 | MATa his3Δ200 lys2-128δ ura3-52 leu2Δ1 chd1Δ::HIS3 spt5-242 |
| GHY611 | MATα his4-912δ lys2-128δ leu2Δ1 trp1D63 SPT5-MYC |
| GHY617 | MATα his4-912δ lys2-128δ leu2Δ1 SPT5-FLAG |
| GHY713 | MATa chd1Δ::URA3 his3Δ200 lys2-128δ leu2Δ1 ura3-52 |
| GHY773 | MATa HA13-CHD1 his3Δ200 lys2-128δ leu2Δ1 ura3-52 |
| GHY1300 | MATα his4-912δ lys2-128δ leu2Δ1 trp1Δ63 HA-SPT6 SPT5-FLAG YPR133C-MYC13::TRP1 |
| KY399 | MATα rtf1Δ100::URA3 leu2Δ1 ura3-52 trp1Δ63 |
| KY620 | MATα rtf1Δ101::LEU2 HA13-CHD1 his4-912δ lys2-128δ leu2Δ1 ura3-52 |
| KY661 | MATa his3Δ200 his4-912δ lys2-128δ leu2Δ1 ura3-52 |
| PJ69-4A | MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ |
| YTT227 | MATa ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 RAD5+ isw1Δ::ADE2 isw2Δ::LEU2 chd1Δ::TRP1 |
The chd1Δ::HIS3 and chd1Δ::URA3 mutations were constructed by a PCR-based method (Woodage et al., 1997). The HA3-CHD1 plasmid pGH204 contains a MunI–HindIII fragment of genomic DNA encompassing CHD1 in which sequences encoding the HA3 tag were inserted by a PCR-based method (Schneider et al., 1996). The presence of the tag inserted immediately after the first ATG codon of Chd1 was confirmed by DNA sequence analysis and immunoblotting. To integrate HA3-CHD1 into the normal CHD1 chromosomal locus, a restriction fragment bearing the full-length HA3-CHD1 gene was transformed into strain GHY713, which carries the chd1Δ::URA3 mutation. Transformants that had replaced the chd1Δ::URA3 allele with HA3-CHD1 were selected by plating to 5-fluoro-orotic acid (5FOA) media and confirmed by PCR and immunoblotting.
Two-hybrid interaction screen
Yeast strain PJ69-4A (James et al., 1996), which contains three different galactose-regulated reporter genes (HIS3, ADE2 and lacZ), was transformed with plasmid pLS28 to tryptophan prototrophy. The resulting strain was then transformed with a yeast genomic DNA library, YL2H-C1, constructed in the Gal4 activation domain-encoding plasmid pGAD-C1 (James et al., 1996). Candidates for Rtf1-interacting proteins were identified by selection on SC-his-trp-leu and SC-ade-trp-leu media, purified, and tested for lacZ expression by a filter assay. From a total pool of ∼540 000 transformants, 43 candidates were chosen for further analysis. Of these, four expressed the reporter genes in a manner dependent on both pLS28 and the library plasmid. DNA sequence analysis of the strongest candidate revealed a fusion between the Gal4 activation domain and amino acids 863–1468 of Chd1.
Identification of CHD1 as a suppressor of spt5Cs–
Previously, we performed a genetic selection for mutations that suppress the cold-sensitive (Cs–) growth defect conferred by the spt5-242 mutation (Hartzog et al., 1998; Squazzo et al., 2002). Suspecting that some suppressor mutations might be SPT2/SIN1 alleles, we crossed a strain carrying the recessive spt2-150 mutation to an spt5-242 mutant. Interestingly, we found that although spt2-150 acted as a recessive suppressor of the Cs– growth defect of spt5-242, the suppression phenotype was not complemented by a SPT2 plasmid. The spt2-150 mutation is a deletion that extends beyond the SPT2 gene (F.Winston, personal communication). By transforming plasmids carrying genes that flank SPT2 into the spt5-242 spt2-150 strain, we found that CHD1 fully complemented the suppression of spt5-242 by spt2-150. When we constructed a new deletion mutation that removed only the ORF of CHD1, we found that it acted as a recessive genetic suppressor of the cold-sensitive growth defect of spt5-242 and that it was completely complemented by a plasmid-borne copy of CHD1. Thus, chd1-null mutations suppress the cold-sensitive growth defect of spt5-242.
Plasmid construction
To construct plasmid pLS28 (GBD-Rtf1), a BamHI restriction site was introduced by oligo-directed mutagenesis immediately 5′ to the ATG codon of RTF1 in plasmid pKA61 (Stolinski et al., 1997). The ∼1.9 kb BamHI–PstI restriction fragment of this plasmid was subcloned into the same sites of pGBT9 (Clontech). The Gal4–Rtf1 fusion protein does not significantly activate transcription of Gal4-dependent genes on its own and partially complements an rtf1Δ mutation.
Mutations in CHD1were constructed in a HA3-CHD1 LEU2 CEN plasmid, pGH277. The mutated HA3-CHD1 gene was then cloned as a PvuII fragment into pRS316. C-terminal deletion mutations were constructed by digesting with PstI, which cuts 3′ of the CHD1 stop codon, plus the appropriate restriction enzyme as indicated by the name of the mutant (Figure 1A). Digested DNA was blunted, gel purified and ligated with OGH34, 5′-TGACTAGCTAGCTAGTCA, a self-complementary oligonucleotide containing stop codons in all three reading frames.
To delete the chromodomains of CHD1, we used PCR mutagenesis to create two AgeI sites flanking the chromodomains of HA3-CHD1. This construct was digested with AgeI, gel purified, and re-ligated to create the chromodomain deletion allele of CHD1. This results in the deletion of amino acids 188–343 of Chd1 and their replacement with a threonine– glycine dipeptide sequence.
To generate the K407R substitution in Chd1, an EcoRI–AvaI fragment from pRS316-HA3-CHD1 was subcloned into pGEM3Zf(+) (Promega). A 75 bp AccI fragment, encompassing the K407 codon, was removed by restriction digestion and replaced with a mutagenic double-stranded 75 bp oligonucleotide to incorporate a single nucleotide change that results in the K407R mutation and creation of a MboII site. The point mutation was confirmed by sequencing and transferred back into pRS316-HA3-CHD1 by gap repair. The final pRS316/CHD1-K407R plasmid was confirmed by restriction digest and by immunoblotting for the HA epitope.
β-galactosidase assays
Transformants of yeast strain PJ69-4A were grown at 30°C in SC-leu-trp media to a density of 1–3 × 107 cells/ml. Extract preparation, β-galactosidase assays and unit calculations were performed as described previously, using the unit definition of OD420/min/mg (Prelich, 1997).
Co-immunoprecipitation and immunoblotting experiments
For Rtf1 co-immunoprecipitations, yeast strain GHY773 was grown at 30°C to a density of 2.5–3.5 × 107 cells/ml in YPD. Cells were harvested and washed in lysis buffer containing 100 mM sodium acetate, 2 mM magnesium acetate, 50 mM Tris–HCl pH 7.4, 10% glycerol, 1 mM dithiothreitol (DTT), 1% NP-40 and protease inhibitors. Co-immunoprecipitations were performed on whole-cell extracts (1.5 mg), prepared by bead beating, as described previously (Hartzog et al., 1998). Pre-immune sera and anti-Rtf1 antisera (Squazzo et al., 2002) were used at a final dilution of 1:3000, and reactions were mixed for 2 h at 4°C and then clarified by centrifugation. Co-immunoprecipitations between Spt5-Flag and HA3-Chd1 were carried out using an affinity-purified rabbit polyclonal anti-HA antibody (Squazzo et al., 2002) or anti-Flag conjugated beads (Sigma), and extracts of cells prepared by grinding frozen cell pellets in a mortar and pestle as described (Lindstrom and Hartzog, 2001; Squazzo et al., 2002). Proteins were separated by SDS–PAGE on 7.5% acrylamide gels and immunoblotted using standard procedures (Harlow and Lane, 1988).
ChIP experiments
Strains were grown at 30°C to a density of 1–2 × 107 cells/ml. For ChIP of Spt5-Flag and HA3-Chd1, strains were grown in YP media. For HA3-Rtf1 and TBP ChIPs, SC-trp media were used to maintain selection for CEN/ARS/TRP1-marked plasmids pLS21-5 and pLS20, which express HA3-tagged Rtf1 or untagged Rtf1, respectively (Stolinski et al., 1997). For ChIP analysis of GAL genes, strains were grown in media containing 2% raffinose and then galactose was added to a final concentration of 2%. To cross-link chromatin, formaldehyde was added to a final concentration of 1%. The cross-linking reaction was quenched after 20 min by addition of glycine. Subsequent steps in the ChIP reactions were performed essentially as described previously (Komarnitsky et al., 2000) except that for TBP, HA3-Rtf1 and HA3-Chd1 ChIPs cell lysis was performed in FA lysis buffer containing 0.5% SDS, and for Spt5-Flag ChIPs this step was performed in 0.1 M Tris pH 8.0, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), followed by dilution into a 6-fold excess of FA buffer. Chromatin was isolated by centrifugation, washed and sheared by sonication to an average size of ∼300 bp. Anti-TBP polyclonal antiserum was a gift of Greg Prelich and was coupled to protein A–Sepharose beads (Fast Flow; Amersham Pharmacia) as described (Harlow and Lane, 1988). Anti-HA antibodies coupled to agarose beads were purchased from Santa Cruz Biotechnology. Antibodies were incubated with sonicated chromatin for 1.5 h (anti-Flag) or overnight (anti-HA) at 4°C. Steps to process the immunoprecipitated chromatin, including the washes, protease treatment, reversal of cross-links and purification of DNA, were performed essentially as described by Kuras and Struhl (1999). Quantitative PCR analysis was performed as follows. For Spt5-Flag ChIPs: 30 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 60 s. For HA3-Rtf1, TBP and HA3-Chd1 ChIPs: 27 cycles of 94°C for 45 s, 55°C for 45 s (GAL10 and PMA1) or 50°C for 45 s (TEF2), and 72°C for 30 s. PCR was carried out in 10 µl reactions using Taq polymerase or Platinum Taq polymerase from Invitrogen, 1 pmol/µl (Spt5-Flag) or 0.25 pmol/µl (HA3-Rtf1, TBP and HA3-Chd1) of each primer and [α-32P]dATP. Samples were separated on native polyacrylamide gels and analyzed on a phosphoimager. PMA1 (Cho et al., 2001) and chromosome VI-R subtelomeric primer sequences (Vogelauer et al., 2000) were described previously. Other primer sequences are available upon request.
Acknowledgments
Acknowledgements
We thank Greg Prelich, Tony Weil, Tim Formosa, Jeff Brodsky and Jennifer Goeckeler for antibodies, Lori Spencer for plasmid construction, Philip James and Martin Schmidt for two-hybrid reagents, and Fred Winston and Toshio Tsukiyama for yeast strains. We are grateful to members of the DeRisi, Buratowski, Struhl, Winston and Laurent laboratories for technical advice. This work was supported by a Mellon Foundation predoctoral fellowship to P.J.C. and grants from the National Institutes of Health to K.M.A. (GM52593 and AI01816), G.A.H. (GM60479) and A.D.J. (GM37049).
References
- Alén C., Kent,N.A., Jones,H.S., O’Sullivan,J., Aranda,A. and Proudfoot,N.J. (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell, 10, 1441–1452. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman,E., Doring,P., Werner,J. and Lis,J.T. (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev., 14, 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram M.J. and Pereira-Smith,O.M. (2001) Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene, 266, 111–121. [DOI] [PubMed] [Google Scholar]
- Bouazoune K., Mitterweger,A., Langst,G., Imhof,A., Akhtar,A., Becker,P.B. and Brehm,A. (2002) The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J., 21, 2430–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois C.F., Kim,Y.K., Churcher,M.J., West,M.J. and Karn,J. (2002) Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol., 22, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.A., Imbalzano,A.N. and Kingston,R.E. (1996) Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev., 10, 1479–1490. [DOI] [PubMed] [Google Scholar]
- Cho E.-J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.S., Lowell,J.E., Jacobson,S.J. and Pillus,L. (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol., 19, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Costa P.J. and Arndt,K.M. (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics, 156, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoucelles C., Pinson,B., Saint-Marc,C. and Daignan-Fornier,B. (2002) Screening the yeast ‘disruptome’ for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem., 277, 27036–27044. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C. (2001) Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene, 275, 19–29. [DOI] [PubMed] [Google Scholar]
- Formosa T., Ruone,S., Adams,M.D., Olsen,A.E., Eriksson,P., Yu,Y., Rhoades,A.R., Kaufman,P.D. and Stillman,D.J. (2002) Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics, 162, 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog G.A., Speer,J.L. and Lindstrom,D.L. (2002) Transcript elongation on a nucleoprotein template. Biochim. Biophys. Acta, 1577, 276–286. [DOI] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.D., Morris,J.R., Wu,C. and Winston,F. (2000) Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D.melanogaster. Genes Dev., 14, 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., Stokes,D.G. and Perry,R.P. (1999) CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma, 108, 10–25. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P., Cho,E.J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J. et al. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol., 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- Lindstrom D.L. and Hartzog,G.A. (2001) Genetic interactions of Spt4–Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics, 159, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D.L., Squazzo,S.L., Muster,N., Burckin,T.A., Wachter,K.C., Emigh,C.A., McCleery,J.A., Yates,J.R.,III and Hartzog,G.A. (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol., 23, 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.L. and Jaehning,J.A. (2002) Ctr9, Rtf1 and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol., 22, 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy,G., Chang,C.H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu,W.H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Hannett,N.M. and Young,R.A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell, 9, 799–809. [DOI] [PubMed] [Google Scholar]
- Prelich G. (1997) Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol., 17, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger,A. and Dye,M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Iyer,V.R., Brown,P.O. and Struhl,K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell, 6, 1297–1307. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schneider B.L., Steiner,B., Seufert,W. and Futcher,A.B. (1996) pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast, 12, 129–134. [DOI] [PubMed] [Google Scholar]
- Shi X., Chang,M., Wolf,A.J., Chang,C.H., Frazer-Abel,A.A., Wade,P.A., Burton,Z.F. and Jaehning,J.A. (1997) Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol., 17, 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S.L., Costa,P.J., Lindstrom,D.L., Kumer,K.E., Simic,R., Jennings,J.L., Link,A.J., Arndt,K.M. and Hartzog,G.A. (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J., 21, 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G. and Perry,R.P. (1995) DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol., 15, 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G., Tartof,K.D. and Perry,R.P. (1996) CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA, 93, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski L.A., Eisenmann,D.M. and Arndt,K.M. (1997) Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 4490–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.G., Steger,D.J., Iyer,V.R. and Johnson,A.D. (2000) The chromo domain protein Chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J., 19, 2323–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer,J., Landel,C.C., Shiloach,J. and Wu,C. (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi,T., Yamaguchi,Y., Watanabe,D. and Handa,H. (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J., 17, 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. (1992) Analysis of SPT genes: a genetic approach towards analysis of TFIID, histones and other transcription factors of yeast. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1271–1293.
- Woodage T., Basrai,M.A., Baxevanis,A.D., Hieter,P. and Collins,F.S. (1997) Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA, 94, 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]