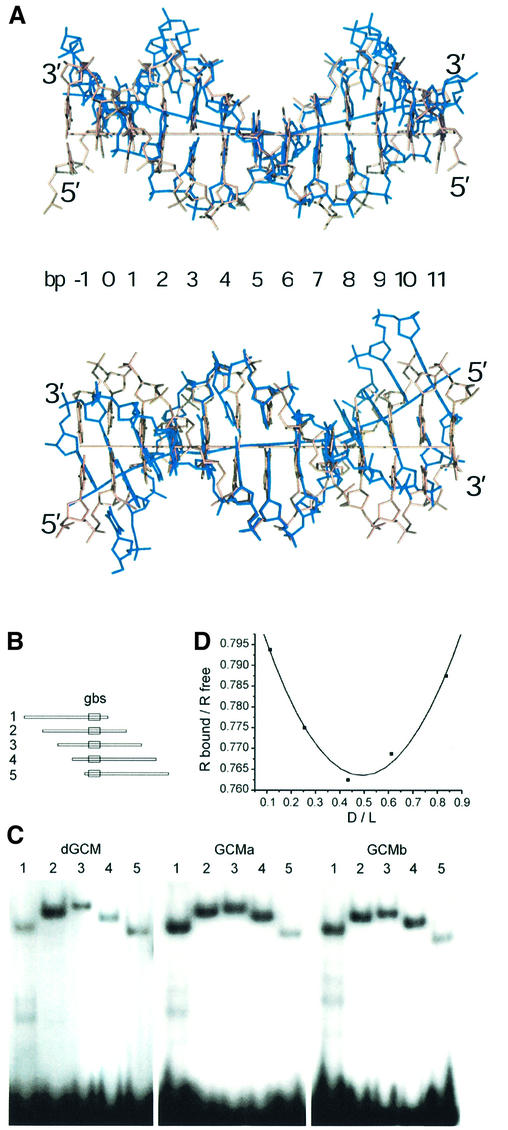
Fig. 4. DNA bending observed in the GCM domain–DNA complex. (A) Two orthogonal views of the 13mer DNA duplex in the GCM domain–DNA complex superimposed with canonical B-form DNA. Strands of the GCM-bound DNA are colored in blue. Helical axes were calculated using the program CURVES (Lavery and Sklenar, 1988). (B) The consensus GCM binding site (gbs) was inserted between the XbaI and SalI sites of pBEND2 (Kim et al., 1989) and retrieved with flanking sequences using the restriction enzymes BglII (1), XhoI (2), XmaI (3), Asp718 (4) and BamHI (5). This generates fragments of identical size with circular permutations of the same sequence and the GCM binding site at varying positions. (C) Circular permutation analyses of DNA bending by electrophoretic mobility shift assays with fragments 1–5 from (A) as probes and the GCM domains of Drosophila GCM (dGCM), mouse GCMa (GCMa) and mouse GCMb (GCMb) expressed in transiently transfected COS cells. (D) Calculation of bending angle for GCMa as described previously (Scaffidi and Bianchi, 2001). The mobility of the protein–DNA complexes (Rbound) was normalized to the mobility of the corresponding free probe (Rfree). The distance of the center of the GCM binding site from the 5′ end of the fragment was divided by the total length of the probe (flexure displacement D/L). The plotted points were interpolated with quadratic functions y = 0.207x2 – 0.203x + 0.813 (r2 = 0.987). The first- and second-order parameters are in close agreement and yield an estimate of 37° for the flexure angle. Similar calculations lead to flexure angles of 34° for Drosophila GCM and 35° for GCMb.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.